Figure 7. DNMT inhibitors can drive Kir4.1 transcription in vitro.
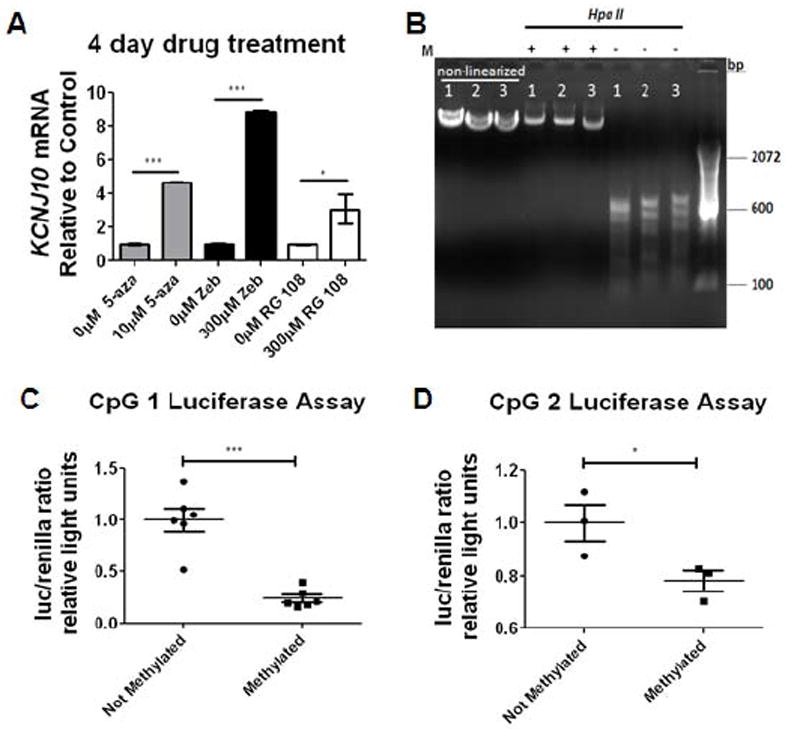
(A) Application of 5-aza, Zebularine (zeb), and RG-108 results in significant increases of Kir4.1 transcription at 4 days following drug treatment. Samples were normalized to gapdh and fold expression is relative to control of each drug (indicated by 0μM). n=3 individual experiments; error bars represent s.e.m.; * (P<0.05); ** (P<0.01) *** (P<0.001). (B) Schematic of methylation of CpG Kir4.1 CpG island is diagrammed. Plasmids were cut releasing Kir4.1 CpG island and then methylated using CpG methylase. Following methylation, CpG island was re-ligated back to non-methylated luc2 (luciferase) plasmid. Methylation (M) of 4.1-CpG 1-3-luc was verified by Hpa II digestion. Hpa II only digests non-methylated DNA. (C, D) Luciferase reporter assay demonstrates that methylation of CpG island 1 and 2 resulted in reduced promoter activity (***p<0.0001 and * p=0.0257; n=6 and n=3). Error bars represent s.e.m.
