Abstract
Plants adapt to their unique soil environments by altering the number and placement of lateral roots post-embryonic. Mutants were identified in Arabidopsis thaliana that exhibit increased lateral root formation. Eight mutants were characterized in detail and were found to have increased lateral root formation due to at least three distinct mechanisms. The causal mutation in one of these mutants was found in the XEG113 gene, recently shown to be involved in plant cell wall biosynthesis. Lateral root primordia initiation is unaltered in this mutant. In contrast, synchronization of lateral root initiation demonstrated that mutation of XEG113 increases the rate at which lateral root primordia develop and emerge to form lateral roots. The effect of the XEG113 mutation was specific to the root system and had no apparent effect on shoot growth. Screening of 17 additional cell wall mutants, altering a myriad of cell wall components, revealed that many (but not all) types of cell wall defects promote lateral root formation. These results suggest that proper cell wall biosynthesis is necessary to constrain lateral root primordia emergence. While previous reports have shown that lateral root emergence is accompanied by active remodelling of cell walls overlying the primordia, this study is the first to demonstrate that alteration of the cell wall is sufficient to promote lateral root formation. Therefore, inherent cell wall properties may play a previously unappreciated role in regulation of root system architecture.
Key words: Cell wall, developmental plasticity, extensin, lateral root, root system development, root system architecture, XEG113.
Introduction
Post-embryonic development is the plant’s means of coping with immobility. It gives plants a way to adapt to the chaotic nature of the world around them. It also allows plants to ‘move’, slowly and methodically, towards that which they need most in life: sunlight, nutrients, and water. The latter two elements reside beneath the soil. Hence, it is the unseen organs of the root system that must respond to environmental cues to forage successfully for nutrients and water.
Hundreds or even thousands of individual roots, branched in a myriad of ways and stretching in nearly all directions, may comprise the root system of a single adult plant. However, what appears to be amazingly complex has a humble origin. For many plants, particularly those belonging to the clade eudicotyledon (including the model plant Arabidopsis thaliana), the complex adult root system originates from a single embryonic root. From this root develop new autonomous lateral roots. These roots give rise to more roots, and so the pattern continues. This process is neither fixed nor random, but rather highly dependent on the environment that surrounds it (Malamy, 2005, 2009; Jung and McCouch, 2013).
Within each root exists the pericycle cell layer, the outermost layer of the plant vascular bundle. The pericycle is the site of origin for new lateral roots (reviewed in De Smet, 2012). In Arabidopsis, the pericycle cells that border the xylem poles are the only ones that are competent to become the founders of new lateral root primordia. The first stage of primordium formation is the anticlinal division of three pairs of adjoining xylem-pole pericycle cells (Dubrovsky et al., 2000; Kurup et al., 2005; De Smet et al., 2006; De Smet, 2012). These cells then undergo a periclinal division to double the number of cell layers in the primordia. Additional rounds of anticlinal and periclinal divisions result in a mature lateral root primordium that is ready to emerge from the confines of the parental root and begin to grow autonomously as a lateral root (Malamy and Benfey, 1997; Peret et al., 2009). During the emergence process, the primordium passes through three overlying cell layers of the parental root: the endodermis, cortex, and epidermis. These cells do not disintegrate, but rather separate from each other to allow the primordium to pass between them (Swarup et al., 2008). Both lateral root initiation and primordia emergence are areas of intensive study and neither is completely understood (i.e. Casimiro et al., 2003; Peret et al., 2009; De Smet, 2012). Both processes are regulated by a combination of intrinsic developmental programmes and responses to environmental cues, such as nutrients, light, and sucrose (Malamy, 2005, 2009; Jung and McCouch, 2013).
Recently, a novel regulator of lateral root emergence was discovered (Swarup et al., 2008). LAX3 is a high-affinity auxin importer expressed in cells overlying lateral root primordia, where its activity regulates expression of putative cell wall remodelling enzymes. Mutants in LAX3 show reduced lateral root emergence that is correlated with a decrease in the expression of cell wall remodelling enzymes. The authors propose that the reduced expression of cell wall remodelling genes may hinder emergence by making it more difficult for cells overlying primordia to separate. Previous studies have also reported expression of putative cell wall remodelling genes around primordia and suggested that the resulting increase in cell wall remodelling proteins allows primordia to pass more easily between overlying cells (Neuteboom et al., 1999; Laskowski et al., 2006; Gonzalez-Carranza et al., 2007). However, knockout of only one of these genes, GLH17, shows a (slight) alteration in lateral root emergence (Swarup et al., 2008).
Here, a genetic screen of mutated Arabidopsis seedlings grown in culture was conducted to identify novel genes that play a role in lateral root primordia development and emergence. Mutants that showed increased lateral root formation underwent a secondary screening process to create functional subcategories of mutations. This lead to the identification of a single mutant where the increased lateral root phenotype was: (1) due to increased emergence; (2) independent of sucrose uptake from the culture media by leaves (previously shown to stimulate emergence; Macgregor et al., 2008); and (3) independent of increased growth of the shoot system, which can indirectly stimulate root system development (Roycewicz and Malamy, 2012). In this mutant, named lateral root development 5 (lrd5), lateral root primordia moved more quickly through all developmental stages, from initiation to emergence of the lateral root primordia from the parent root. lrd5 has a defect in Xyloglucotransferase113 (XEG113), a gene recently shown to play a role in the arabinosylation of cell wall extensins (Gille et al., 2009). Examination of the roles of closely related genes, as well as unrelated genes known to be involved in various aspects of cell wall formation, revealed that many but not all alterations in cell wall formation lead to dramatic increases in lateral root formation. These findings demonstrate that lateral root primordia emergence is a default process that is constrained by properly synthesized cell walls and suggest that inherent cell wall properties may play a previously unappreciated role in regulation of root system architecture.
Materials and methods
Plant growth conditions
Seeds were surface sterilized in 100% bleach plus Tween-20 for 3min while vortexing and rinsed three times using autoclaved and filter-sterilized water. Seeds were imbibed for 2 or more days at 4 °C. Seeds were planted on agar media (described below) approximately 2cm from the top edge of a 100×100cm square Petri plate, with nine seeds per plate. Plates were wrapped with parafilm and placed vertically in a growth chamber with a 16/8 light/dark cycle (50–60 μmol) at 22 °C. Plants were grown for 15 d unless mentioned otherwise.
To isolate shoot tissues from contacting agar media, a strip of parafilm (about 2×9cm) was placed at the top of a Petri plate and seeds were sown just below the parafilm. Parafilm was sterilized for at least 16h in 95% ethanol.
Media composition
Control media contained the following components (per litre): 10g sucrose, 0.5g MES, 100ml Murashige and Skoog basal salt micronutrient solution (10×, M0529; Sigma Aldrich), 5ml of 1M KNO3, 5ml of 1M NH4NO3, 0.33g CaCl2·6H20, 0.1807g MgSO4, 0.17g KH2PO4, and 7.0g agar. Media was brought to pH 5.7 using 1M KOH prior to addition of agar, and autoclaved for 30–45min.
Other media was made by supplementing the control media with varying amounts of mannitol, sucrose, or equimolar amounts of KNO3 and NH4NO3. Repressive media was made by supplementing the control media with (per litre) 35g sucrose, 15ml of 1M KNO3, and 15ml of 1M NH4NO3.
Determination of total lateral root length
Digital images of plant roots (taken using a Canon SD1000 digital camera) were traced by hand using ImageJ. For each seedling, the length of all lateral roots was summed to give the total lateral root length.
Microscopic analysis of primordia initiation and emergence
Seedling roots were cleared by incubating samples sequentially in the following solutions: (1) 15min in 20% methanol acidified with 4% concentrated hydrochloric acid at 55 °C; (2) 15min in 7% NaOH in 60% ethanol at room temperature; (3) 10min in 40% ethanol at room temperature; (4) 10min in 20% ethanol at room temperature; and (5) 10min in 10% ethanol at room temperature. Glycerol (50%) was added to the last solution and stored until mounted on glass slides. Samples were observed using DIC optics on a Leica DMR microscope. Primordia were scored according to the staging system developed by Malamy and Benfey (1997).
Plant material
Original mutant screen was performed with seeds from the Feldmann T-DNA collection (Feldmann, 1991) obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Approximately 100 pools containing 10 lines each (ABRC CS84441) were screened. Germplasm containing the pIAA14::mIAA14-GR construct was a gift from Hidehiro Fukaki. The following mutants were obtained from the Arabidopsis Biological Resource Center: mur1-1 (CS6243), mur2-1 (CS8565), mur3-1 (CS8566), mur4-1 (CS8568), mur5-1 (CS8572), mur6-1 (CS8573), mur7-1 (CS8574), mur8-1 (CS8575), mur9-1 (CS8576), mur10-1 (CS8577), mur11-1 (CS8579), prc1-1 (CS297), ixr1-1 (CS18), xxt1/xxt2 (CS16349), SALK_066991, SALK_053158, SALK_058092, rra1 (CS825155), and rra2 (CS803312).
Determination of shoot size
Shoot size was estimated by cutting seedlings at the root–shoot junction and transferring aerial tissues to a 1.5-ml tube containing 0.5ml ethanol and incubating for 15h to extract chlorophyll. Ethanol solution (0.2ml) from each sample was transferred to 96-well plates. The absorption of the samples at 430nm was analysed using a plate reader (Tecan Safire II). To validate this method, plants were grown for 7–16 d on control media or on control media supplemented with 162mM mannitol to create plants of varying sizes. For each condition and age, 5–15 seedlings were cut at the root–shoot junction, aerial tissues were pooled, and their mass was measured. The same tissue was then extracted with ethanol and A 430 and mass were compared. There was an excellent correlation between A 430 and mass for plants of all ages grown under multiple conditions (Supplementary Fig. S2 available at JXB online). All data points represent the average on a per-plant basis of the pooled samples.
Cloning of causal mutations
To identify the location of T-DNA insertions within the genomes of all mutants, thermal asymmetric interlaced PCR (TAIL-PCR) was performed on genomic DNA using the method and nonspecific primer sequences previously described by Liu et al. (1995) and primers within the T-DNA. The resulting PCR products were cloned into the vector PCR4 using the TOPO cloning system (Invitrogen) and sequenced.
Creation of phylogeny
Phylogenetic analysis was performed using AlignX, which is part of the VectorNTI 10 software package. This program uses the neighbour-joining algorithm to construct unrooted phylogenies of the provided sequences. Amino acid sequences of LRD5 and several homologous sequences were used to create a phylogenetic tree, with a distantly related homologue from Ostreococcus lucimarinus as an outgroup.
Tracking primordia emergence using pIAA14::mIAA14-GR construct
Seedlings containing the pIAA14::mIAA14-GR construct in both the lrd5-2 and wild-type backgrounds were grown on repressive media supplemented with dexamethasone (final concentration 1 μM) for 9 d. After that time, seedlings were transferred to repressive media without dexamethasone (some seedlings were cleared and inspected to confirm that no lateral root primordia initiated in these seedlings). After transfer, seedlings were allowed to grow for 5 d. At that point, seedlings were cleared and analysed for the stage of each primordium according to the staging described by Malamy and Benfey (1997).
Construction of GFP constructs
A PCR8 vector containing the cDNA sequence of LRD5 was kindly provided by Sascha Gille and Markus Pauly. This vector was recombined with the vector pMDC83 (Curtis and Grossniklaus, 2003) using the Gateway LR Clonase Enzyme Mix (Invitrogen). The resulting vector was digested with BamHI and PstI to remove the 35S promoter, and 2kb of genomic sequence immediately upstream of the LRD5 start site was ligated in its place. The final vector was transformed into lrd5-2 via Agrobacterium-mediated transformation. Eight independent lines were selected on agar plates containing hygromycin. Of these, two lines showed strong GFP expression and only these two complemented the mutant phenotype in lrd5-2.
Results
Secondary screens identify distinct classes of lateral root mutants
Increased formation of lateral roots is a commonly reported phenotype in Arabidopsis mutants. Therefore, it is useful to have a pipeline to categorize mutants into subgroups representing known mechanisms for increased lateral root formation. Such a pipeline is described here for a group of eight mutants that were isolated with increased lateral root formation, and which were rescreened for lateral root emergence mutants of interest.
The mutant screen used conditions demonstrated in Deak and Malamy (2005) and Macgregor et al. (2008) to strongly repress lateral root formation. For this laboratory assay, repressive conditions were defined as 1× MS basal salts with 20mM each KNO3 and NH4NO3, 4.5% sucrose, and low light (50–60 μmol). Seedlings grown under these repressive conditions have been previously reported to show a dramatic decrease in lateral root formation compared with seedlings grown under control conditions (control conditions were defined as 1× MS basal salts, 5mM each KNO3 and NH4NO3 and 1% sucrose, and low light (50–60 μmol) (Macgregor et al., 2008). Seedlings from 100 pools of T-DNA mutagenized lines in the Wassilewskija (Ws) background (Feldmann, 1991; ABRC CS84441) were screened. This work identified eight mutants that showed a robust increase in lateral root formation on repressive conditions as compared to the wild type (Ws; see Fig. 1A for examples).
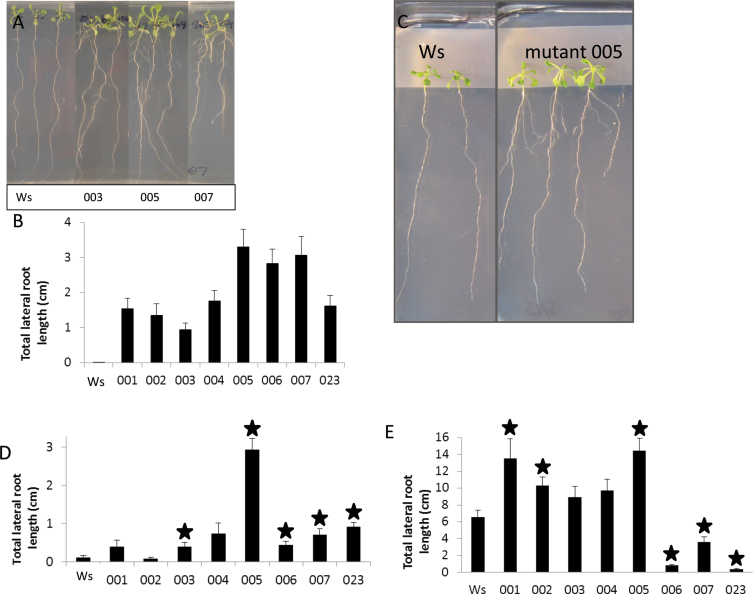
Fig. 1.
Characterization of eight mutants with increased lateral root formation compared to the wild type (Ws). (A) Three mutants showing increased lateral root formation after 15 d of growth on repressive conditions. (B) Total lateral root length for eight mutant lines compared to the wild type grown on repressive media for 15 d: all show a statistically significant increase compared to the wild type (P<0.01, Student’s paired t-test). (C) Mutant grown on parafilm to prevent contact between aerial tissues and media, continuing to show the lateral root phenotype. (D) Total lateral root length for eight mutant lines grown on repressive media for 18 d with aerial tissue isolated from media by parafilm; stars indicate statistically significant increases compared to the wild type (P<0.05, Student’s paired t-test). (E) Total lateral root length for eight mutant lines grown on control media for 14 d: mutants 001–005 show an increase in lateral root formation, mutants 006, 007 and 023 show a decrease in lateral root formation; stars indicate statistically significant differences compared to the wild type (P<0.05, Student’s paired t-test). Data are mean±standard error (n= 22–34).
Lateral root formation is repressed under these screening conditions due to a number of factors. One factor is that osmotic potential affects the permeability of the aerial tissues to sucrose in the media, with lower osmotic potential reducing sucrose uptake and thereby reducing lateral root formation (Macgregor et al., 2008). Another factor is osmotic repression of the overall growth rate of the plant or of the shoot system, which may exert an indirect effect on growth of the root system (Roycewicz and Malamy, 2012). Additionally, high levels of nitrate in the media have a repressive effect on lateral root formation that is independent of osmolarity, shoot growth, or shoot permeability to media sucrose (Roycewicz and Malamy, 2012). Based on this information, it is possible to predict five types of mechanisms that would allow a mutant to exhibit an increase in lateral root formation in this assay system: (1) increased permeability to media sucrose at the shoot, as has previously been shown for mutants in cuticle formation (Macgregor et al., 2008); (2) constitutive increase in shoot system or whole plant growth rates; (3) reduction in nitrate-mediated repression of lateral root formation; (4) specific increase in lateral root formation independent of shoot growth; and (5) some other process is altered in a way that has yet to be uncovered. Mutants of type 4 would most likely lead to a greater understanding of the lateral root emergence process, and isolating such mutants was therefore the goal of this study.
To characterize the mutants, the average total length of all lateral roots was first determined for each line. As expected, each of the eight mutant lines identified showed a statistically significant increase in total lateral root length compared to wild type (Ws) when grown under repressive conditions for 15 d (Fig. 1A, B).
To identify type 1 mutants that overcame repression of lateral root formation via an increase in sucrose uptake by the shoot, this study investigated whether each mutant still showed a lateral root phenotype under repressive conditions with parafilm blocking aerial tissue contact with media sucrose, as in Macgregor et al. (2008). Five out of the eight mutants still showed a significant and reproducible increase in lateral root formation when shoots were isolated on parafilm (Fig. 1C, D), indicating that their phenotype did not depend on sucrose uptake from the media. In contrast, it is likely that mutants 001, 002, and 004 had increased lateral root formation due to increased sucrose uptake at the leaves.
Next, all eight mutants were grown under control conditions to see if any of the mutants had a growth condition-specific phenotype (i.e. ability to overcome low osmotic potential or high nitrate repression). Wild-type (Ws) seedlings grown under control conditions formed numerous lateral roots at 2 weeks of age. Interestingly, the eight mutant lines separated into two groups when grown under control conditions (Fig. 1E). Five mutants (001–005) showed an increase in lateral root formation compared to Ws under control conditions (although due to the high variability of lateral root formation on this condition, only three could be confirmed as being significantly different). Mutants 001, 002, and 004 were suggested to have increased sucrose uptake from the media under low osmotic potential (type 1), and increased lateral root formation under all conditions is consistent with that idea. Interestingly, mutants 006, 007, and 023 grown on control media showed a significant decrease in lateral root formation compared to wild type (Ws). Opposite phenotypes on the two growth conditions is not predicted for mutants of types 2, 3, or 4, and therefore these three mutants must have defects in a novel, environmentally responsive regulatory mechanism (type 5).
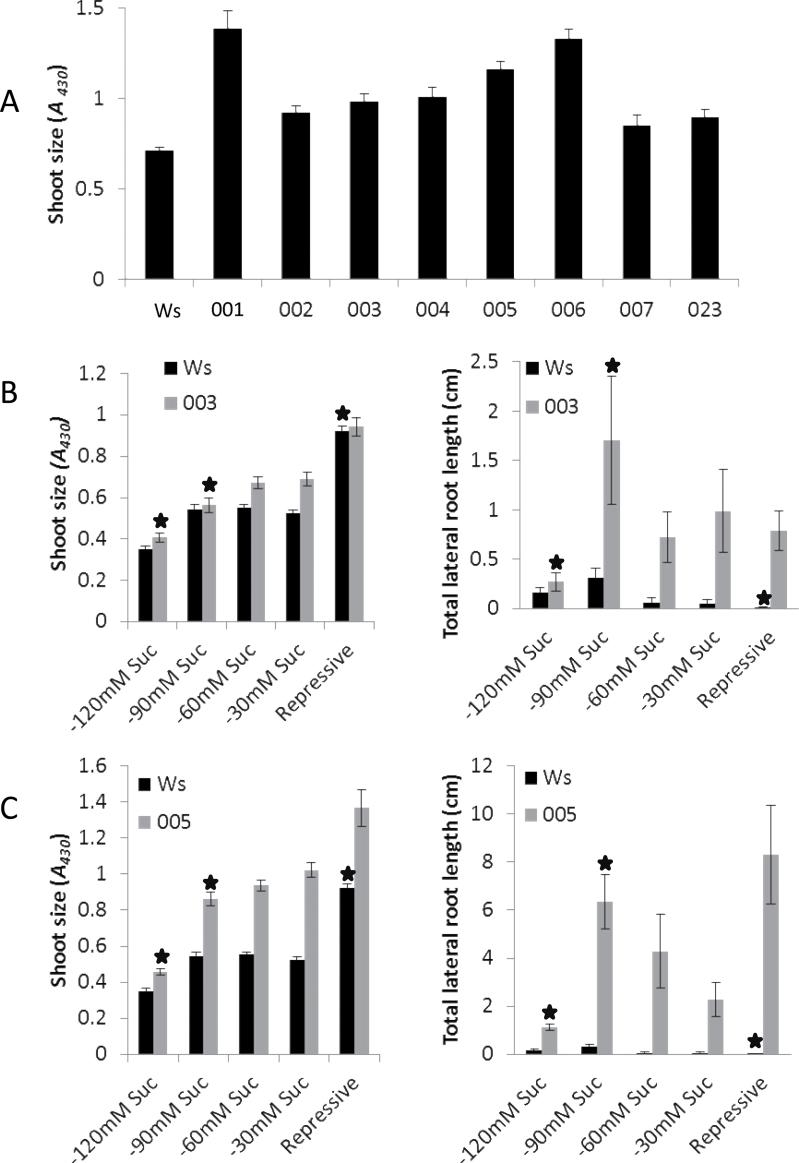
Mutants 003 and 005 became the focus of further investigation, as they had the potential to be type 4 mutants (defects specific to the lateral root formation process). To determine whether they were indeed type 4 or were exhibiting lateral root phenotypes due to increases in shoot or overall plant growth rates (type 2), shoot systems were examined. Both mutants 003 and 005 (as well as all others mutants described here) appeared to have an increased shoot size when examined visually (Fig. 1A and Supplementary Fig. S1 available at JXB online). To confirm this observation, seedlings were quantified for chlorophyll content using A 430 as a proxy for shoot size (see Methods and Supplementary Fig. S2 available at JXB online). All mutants exhibited a significant increase in shoot size (Fig. 2A). To test whether root and shoot system sizes in mutants 003 and 005 are always correlated, or whether an increase in lateral root formation per unit shoot size could be found, mutants were grown on media with varying amounts of sucrose. Reducing the sucrose content in the culture media modulates the size of aerial tissues. In each medium, sucrose was replaced with an equivalent amount of the non-metabolizable sugar mannitol (to maintain a constant osmotic potential). For both mutants, reducing the concentration of sucrose resulted in seedlings with smaller or similar shoot sizes compared to Ws seedlings grown under repressive conditions (Fig. 2B, C, right panels). However, even when manipulation of the media lead to mutant plants with a smaller shoot size than Ws, the mutants still exhibited a higher total lateral root length than the Ws seedlings on repressive media (Fig. 2B, C, left panels). These results confirm that both mutants 003 and 005 are capable of forming more lateral roots per unit shoot size. Type 2 mutants are predicted to have a correlated increase in shoot and root growth, either due to an overall increase in the whole plant growth rate or an indirect effect of increased shoot growth on root growth. Thus 003 and 005 are not type 2 mutants, but rather demonstrate a root-specific phenotype, as predicted for type 4 mutants.
Fig. 2.
Shoot size and total lateral root length for mutants and the wild type (Ws) grown on repressive media and media with reduced sucrose and compensating mannitol content. (A) Shoot size for eight mutant lines compared to the wild type grown on repressive media for 15 d: all show a statistically significant increase compared to Ws (P<0.01, Student’s paired t-test). (B) Mutant 003 shows a significantly reduced shoot size (left panel) and a significantly increased total lateral root length (right panel) at 120 and 90mM sucrose when compared to the wild type (stars, P<0.01, Student’s paired t-test). (C) Mutant 005 shows a significantly reduced shoot size at 120mM sucrose (right panel) and significantly increased total lateral root length (left panel) (stars, P<0.01, Student’s paired t-test) compared to the wild type. Data are mean±standard error (n=16–35).
Mutation in At2g35610 is responsible for lateral root phenotype in mutant 005 (lrd5-1)
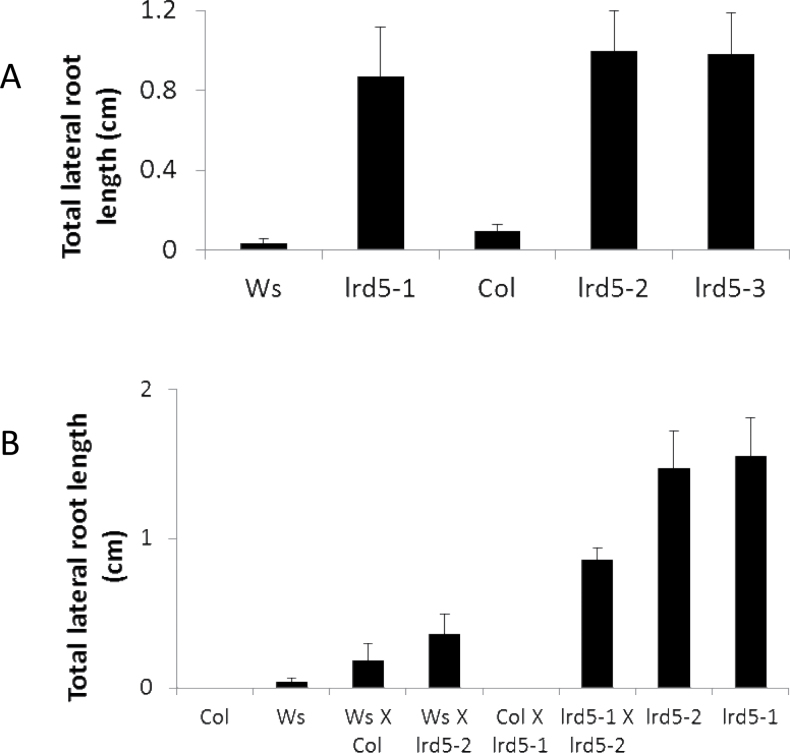
Thermal asymmetric interlaced PCR was employed to identify the genomic location of the T-DNA insertion in mutants 003 and 005. No T-DNA was found in the 003 line either by PCR or by Southern blot analysis (not shown). However, two genomic sequences were obtained from TAIL-PCR of mutant 005, corresponding to At2g35610 and At1g20640. Segregation analysis was performed in order to determine whether either of the T-DNA insertions identified cosegregated with the mutant phenotype. The mutant was crossed to the parental background (Ws) and the resulting F1 progeny were allowed to self-fertilize. None of the F1 progeny showed a mutant phenotype, indicating that the mutation was recessive. The F2 progeny were screened for mutant phenotype, and were tested via PCR using primers specific for each T-DNA insertion (one primer internal to the T-DNA and another in the genomic DNA). Surprisingly, both T-DNA insertion sites cosegregated perfectly with the mutant phenotype.
Mutant 005 was designated lateral root development 5 (lrd5-1). Of the two T-DNA insertions identified in lrd5-1, the first was located within an intron in gene At2g35610 and the second within an exon in gene At3g20640. Since the T-DNA insertions were predicted to be on separate chromosomes, the fact that both T-DNA insertions cosegregated perfectly with each other and with the mutant phenotype suggests the possibility of genomic rearrangement in lrd5-1. To determine which mutation was responsible for the mutant phenotype, SALK T-DNA alleles of At2g35610 SALK_066991 (lrd5-2) and SALK_058092 (lrd5-3) were examined (no alleles of At3g20640 were available at the time of this work). These alleles are in the Col background. Both lrd5-2 and lrd5-3 showed increased total lateral root length, like the original mutant lrd5-1, when grown with parafilm blocking contact between the aerial tissues and media (Fig. 3A). Additionally, a cross between the original T-DNA mutant lrd5-1 and lrd5-2 failed to rescue the mutant phenotype (Fig. 3B; since the two alleles are in different backgrounds, crosses are included to eliminate the possibility of interactions between mutant alleles and disparate genetic backgrounds) These results confirm that the T-DNA insertion in At2g35610 is causal for the mutant phenotype in lrd5-1.
Fig. 3.
Phenotyping and complementation tests with three alleles of LRD5. (A) Total lateral root length for several alleles of lrd5 grown with parafilm isolating aerial tissue from media: all mutants are significantly different from their respective wild-type backgrounds (P<0.01, Student’s paired t-test); data are mean±standard error (n=23–63); lrd5-1 is in the Ws background while lrd5-2 and lrd5-3 are in the Col background. (B) Total lateral root length for the wild type, two alleles of lrd5, F1 plants from complementation test crosses, and F1 plants from control crosses; data are mean±standard error (n=17–53). Differences between lrd5-1×lrd5-2 and all controls are significant (P<0.01, Student’s paired t-test).
LRD5 modifies cell wall proteins
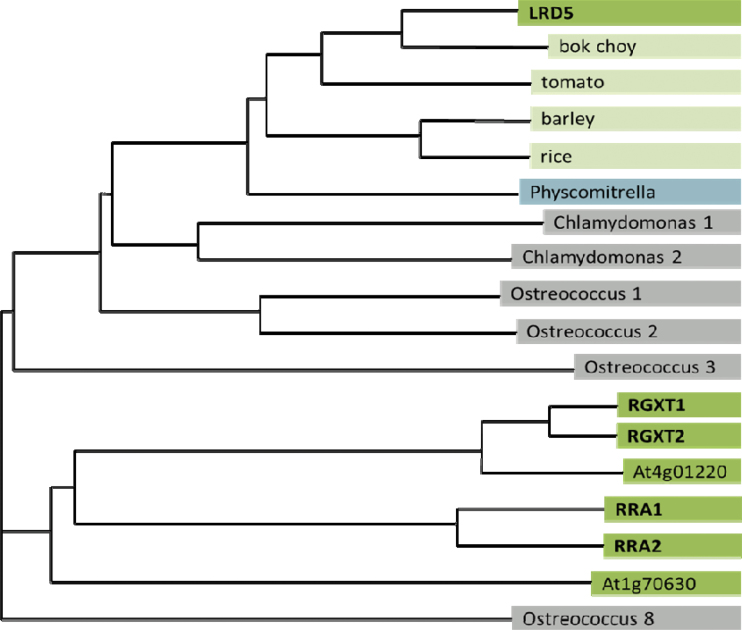
LRD5 encodes a gene with no known domains or motifs. Previous computational studies have grouped LRD5 within the glycosyltransferase family 77 based on homology to other glycosyltransferases (www.cazy.org). This result was confirmed by a BLAST search using the predicted protein sequence of LRD5 against the NCBI sequence database. Six Arabidopsis genes showed significant homology to LRD5 (Fig. 4), all of which belong to the glycosyltransferase family 77 (www.cazy.org). Several of these genes have been previously identified to play a role in cell wall biosynthesis (Egelund et al., 2006, 2007, 2008). The most closely related gene, At1g70630, is predicted to encode a protein with 51% amino acid similarity over a stretch of 155 amino acids located towards the middle of the protein sequence. However, no significant similarity was found along the rest of the protein sequence. This gene was recently named Reduced Arabinose Yariv1 (RAY1) and shown to play a role in arabinosylation of cell wall arabinogalactan proteins (AGPs; Gille et al., 2013).
Fig. 4.
Amino acid phylogeny using the neighbour-joining method using select sequences identified from a BLAST search using the protein sequence of LRD5. Several species included several homologous sequences, which are individually numbered. RGXT1, RGXT2, RRA1, and RRA2 have all been shown to be involved in cell wall biosynthesis.
The BLAST search also identified sequences in over 30 plants, the moss Physcomitrella patens, and three algae (O. lucimarinus, Ostreococcus taurii, and Chlamydomonas reinhardtii) that showed greater homology to LRD5 than the closest homologue in Arabidopsis (selected examples shown in Fig. 4). Given that Arabidopsis diverged from Ostreococcus and Chlamydomonas roughly 1 billion years ago (Yoon et al., 2004), this suggests that LRD5 diverged from its closest Arabidopsis homologue at least that long ago.
Recently, mutants in LRD5 (lrd5-2/xeg113-2 and lrd5-3/xeg113-3) were characterized and shown to have an increase in etiolated hypocotyl length, rosette size, and early inflorescence bolting (Gille et al., 2009) and root hair growth (Velasquez et al., 2011). These mutants contained reduced arabinosylation of cell wall extensins and the authors proposed that LRD5 functions as an extensin arabinosyltransferase. The authors concluded that extensin glycosylation by At2g35610 is an important determinant for proper cell elongation. However, it was not immediately obvious how these results relate to the lateral root phenotype seen in lrd5 mutants.
Mutants in LRD5/XEG113 show an increased rate of lateral root development and emergence
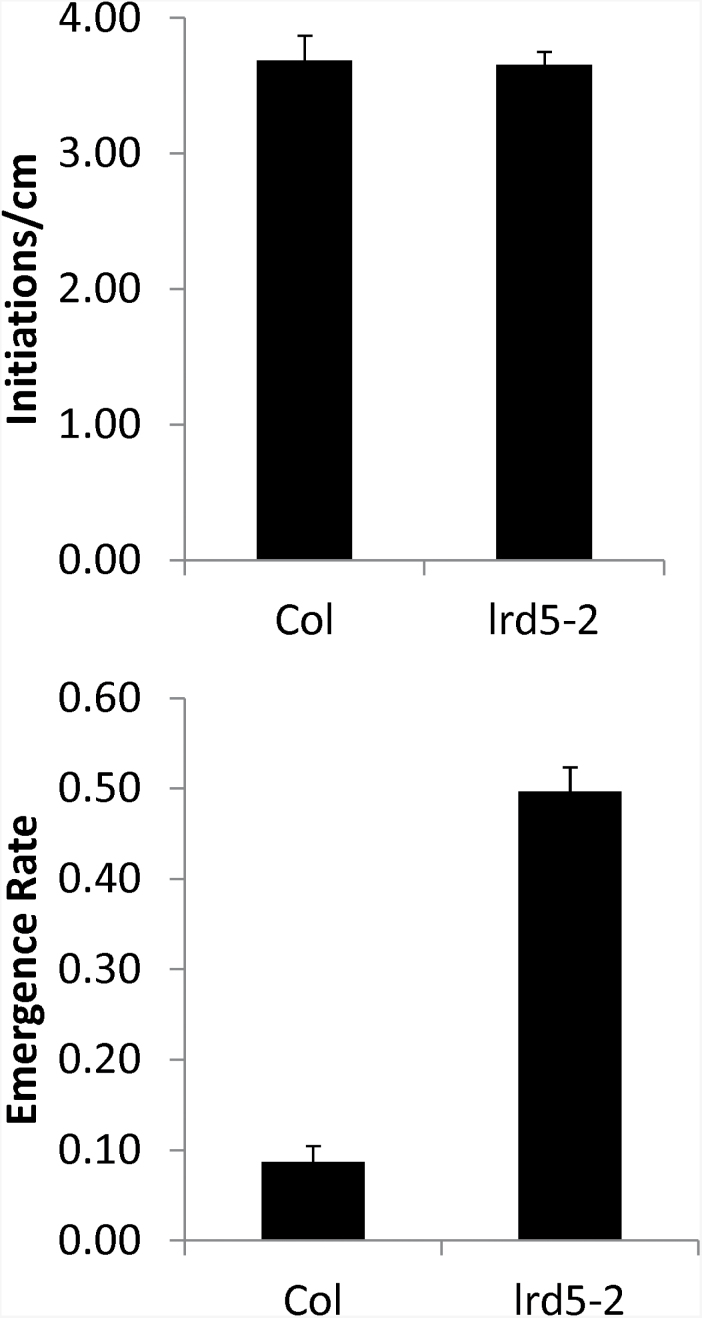
The increase in lateral root formation observed in mutants in LRD5/XEG113 may be due to an increase in the number of initiated primordia, the rate at which primordia emerge, or both. This question was addressed by analysing the density of initiations and the frequency of emergence in 14-day-old lrd5-2 and wild-type (Col) seedlings. lrd5-2 seedlings showed no increase in the number of primordia, but a large increase in the frequency with which these primordia emerge (Fig. 5). (A striking increase in emergence was also seen in lrd5-1 in comparison to its wild-type background (Ws) (Supplementary Fig. S3A available at JXB online), although the phenotype was less pronounced due to the higher emergence rate in Ws vs. Col). These results suggest that aberration in some aspect of the emergence process strongly contributes to the lateral root formation phenotype in lrd5 mutants, although the possibility remains that an increase in lateral root growth rate also contributes to the phenotype.
Fig. 5.
Number of initiations and emergence rate for lrd5-2 and the wild type (Col). Emergence rate is measured as the number of emerged primordia divided by total number of primordia. A total of 161 primordia were analysed for lrd5-2 and 254 for Col. No difference in initiations per cm was seen in the mutant (Student’s paired t-test). In contrast, emergence rate was significantly increased (P<0.01, pooled z-statistic). Data are mean±standard error (161 primordia in 10 seedlings lrd5-2 and 254 primordia in 10 seedlings for Col).
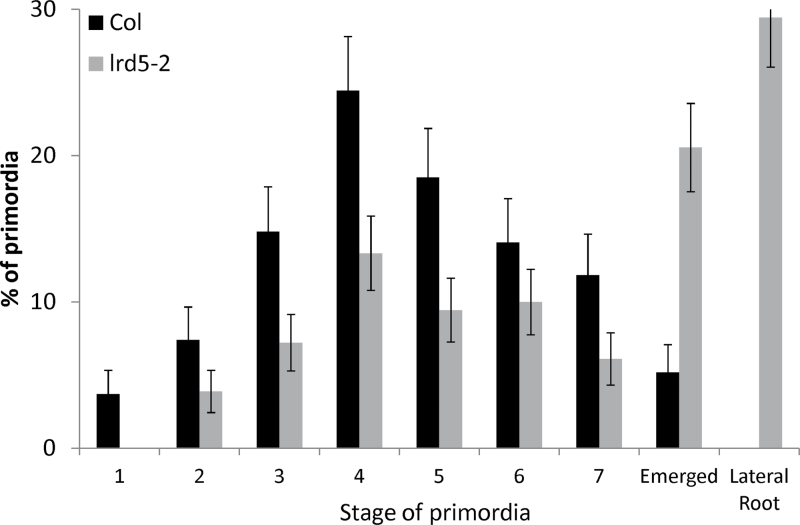
Given that primordia emerge more frequently in lrd5, the developmental stages of the primordia were next examined. To more accurately monitor the developmental process of lateral root primordia in wild-type and lrd5 seedlings and to allow a detailed comparative time course of lateral root development, lateral root initiation was synchronized. lrd5-2 was crossed into plants containing the pIAA14::mIAA14-GR, a dexamethasone-inducible dominant allele of IAA14 (Fukaki et al., 2005). IAA14 (SLR1) is a major component of auxin-mediated lateral root initiation, and a dominant mutation of IAA14, mIAA14, completely abolishes lateral root initiation. Grown in the presence of dexamethasone, pIAA14::mIAA14-GR seedlings do not initiate any primordia; upon removal of dexamethasone, pIAA14::mIAA14-GR seedlings initiate primordia in young tissues. This system thus allows for the synchronous initiation of primordia in seedlings old enough to form lateral roots. Wild-type (Col) and lrd5-2 seedlings containing the pIAA14::mIAA14-GR construct were grown on repressive media containing dexamethasone for 9 d and then transferred to media lacking dexamethasone. After 5 d, the seedlings were cleared and primordia were scored based on the staging described by Malamy and Benfey (1997) (Fig. 6). lrd5-2/pIAA14::mIAA14-GR seedlings contained fewer primordia than the wild type at each of the seven pre-emergence stages, but more emerged primordia. Since no differences in initiation rate are predicted in lrd5-2, this result must be explained by an increase in the rate at which primordia progress through the entire developmental process. The reduction even in stage 1 and 2 primordia indicates that the increased rate of development starts around the time that primordia first initiate.
Fig. 6.
Proportion of lateral root primordia at each stage 5 d after release of SLR1 inhibition of initiation in lrd5-2 and wild type (Col). All comparisons except stage 6 are statistically significant (P<0.05, pooled z-statistic. Data are mean±standard error (n=16).
LRD5/XEG113 does not directly affect whole-plant growth
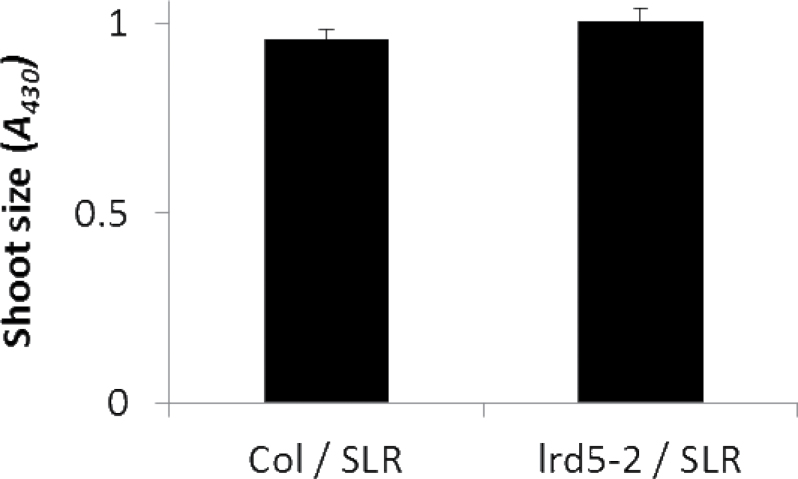
The increase in lateral root formation per unit shoot size (Fig. 2) suggests that the effects of the LRD5/XEG113 mutation are root specific. This would further predict that the increase in aerial tissue growth observed here (Fig. 1) and by Gille et al. (2009) in lrd5/xeg113 mutants is a result of the increase in lateral root growth rates. To test this idea, wild-type (Col) and lrd5-2 seedlings containing the pIAA14::mIAA14-GR construct were grown on media with dexamethasone for 14 d. Indeed, when lateral root formation was inhibited in lrd5-2/xeg113, shoot size was indistinguishable from the wild type (Fig. 7). This indicates that changes in shoot growth in lrd5 are secondary, and not causal, to increased lateral root formation.
Fig. 7.
Aerial tissue size (A 430) of the wild type and lrd5-2 with mIAA14 induced by dexamethasone. Differences between the two genotypes are not statistically significant (Student’s paired t-test). Data are mean±standard error (n=39).
LRD5/XEG113 is expressed in trichoblasts and cells near the root tip
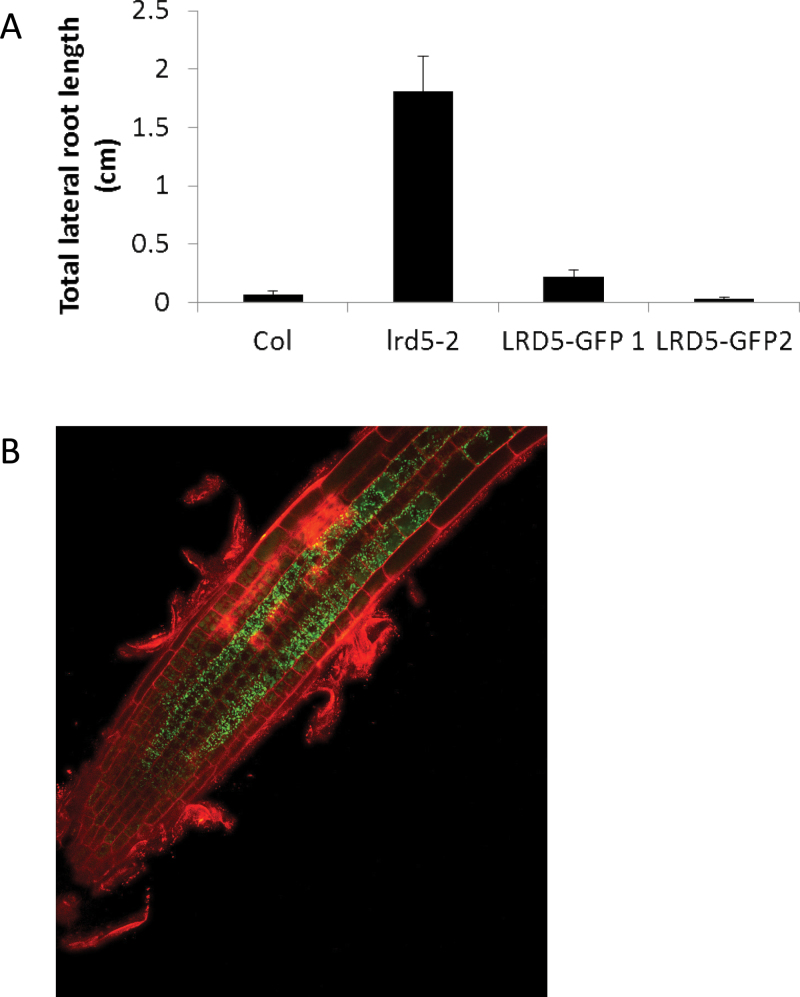
To better understand how XEG113/LRD5 functions in the root to affect lateral root development, a transgenic construct consisting of 2kb promoter sequence driving the wild-type cDNA fused to GFP (pLRD5::LRD5-GFP) was created. This construct was transformed into lrd5-2 and two independent lines were identified that showed strong GFP expression and also rescued the mutant lateral root phenotype by reducing total lateral root length (Fig. 8A) and lateral root emergence rates (Supplementary Fig. S3B available at JXB online).
Fig. 8.
Endogenous expression pattern of LRD5. (A) Total lateral root length for lrd5-2 and lrd5-2 with two separate rescue constructs. lrd5-2 is significantly different from all other genotypes (P<0.01, Student’s paired t-test). Data are mean±standard error (n=47–77). (B) Strong GFP expression is seen in the trichoblast cell file in plants transformed with constructs shown in A.
LRD5/XEG113 is most strongly expressed in the trichoblast cell file just basal to the primary root tip, with decreasing expression intensity in more mature regions of the root (Fig. 8B). These results are consistent with the publicly available microarray data (www.arexdb.org). The microarray data also suggests that LRD5 is expressed at low levels in most cell layers just behind the primary root tip. This expression is barely visible using fluorescence microscopy (not shown). No expression is predicted or can be seen in lateral root primordia or cells overlying primordia (not shown).
The finding that LRD5/XEG113 is expressed near the root tip and not expressed in or around primordia suggests that this gene either acts non-cell autonomously or plays a role in early wall formation in cells that will later play a role in primordia emergence (i.e. cells that will come to overly a primordium later in development). Since it is difficult to imagine how trichoblast cell wall composition would affect lateral root primordia emergence, it is hypothesized that the second model is correct (see Discussion).
Related and unrelated cell wall mutants also show a lateral root phenotype
Mutations in LRD5/XEG113 result in reduced arabinosylation of extensins in the cell wall (Gille et al., 2009) and increased rate of lateral root development and emergence (this study). To test whether other Family 77 glycosyltransferases showed lateral root phenotypes under the screening condition, knockout lines were obtained for At1g70630 (RAY1, the closest homologue to XGE113/LRD5 in Arabidopsis), RRA1 and RRA2. The latter two mutants, although altered in closely related and tandemly duplicated genes, show slight reductions in arabinose content (Egelund et al., 2007). (Mutants in RGXT1, RGXT2, and RGXT3, previously studied Family 77 genes, were not tested as RGXT1 and RGXT2 are tandemly duplicated genes with no detectible phenotypes/defects when mutated and confirmed alleles of RGXT3 are not available). Whereas rra1 and rra2 had no detectible lateral root phenotype, the mutant allele of At1g70630 (Salk_053158, ray1) showed a dramatic increase in lateral root formation compared to the wild type (Col) (Table 1 and Fig. 9). To see if At2g35610 and At1g70630 act in parallel, lrd5-2/xeg113 and ray1 were crossed to form a double mutant. Although the double mutant did not have altered lateral root formation compared to ray1 (the mutant with most lateral root formation) with aerial tissues contacting media, on parafilm the double mutant showed a much stronger phenotype compared to both lrd5-2 and ray1 (Table 1). RAY1 was recently shown to play a role in arabinosylation of AGPs and not extensins (Gille et al., 2013). This suggests that two Family 77 glycosidases with varying functions in cell wall modification affect lateral root formation and that the lrd5/xeg113 phenotype is not specific to extensin modification.
Table 1.
Total lateral root length for cell wall mutants grown with and without parafilm to isolate shoots from mediaValues are mean±standard error. Asterisks indicate statistically significant deviations from wild type (P<0.05 using Student’s paired t-test). Total lateral root length for lrd5-2 and ray1 are significantly different from the lrd5-2/ray1 double mutant. AGP, arabinogalactan protein; NK, not known.
| Perturbed in: | Total lateral root length (cm) | |||
|---|---|---|---|---|
| Gene | Cell wall | Off parafilm | On parafilm | |
| Col | NK | NK | 0.18±0.08 | 0.10±0.05 |
| Cell wall mutants | ||||
| mur1 a | At3g51160 | Fucose | 2.27±0.24* | 1.65±0.26* |
| mur2 a | At2g03220 | Fucose | 0.01±0.00 | – |
| mur3 a | At2g20370 | Fucose | 0.41±0.16* | 0.24±0.15 |
| mur4 a | At1g30620 | Arabinose | 0.86±0.12* | 2.40±0.35* |
| mur5 a | NK | Arabinose | 0.13±0.05 | – |
| mur6 a | NK | Arabinose | 0.09±0.03 | – |
| mur7 a | NK | Arabinose | 0.05±0.02 | – |
| mur8 a | NK | Rhamnose | 1.23±0.19* | 0.73±0.34* |
| mur9 a | NK | Fucose, xylose | 2.94±0.28* | 0.09±0.04 |
| mur10 a | At5g17420 | Cellulose, fucose, arabinose, xylose | 0.11±0.03 | – |
| mur11 a,b | At3g59770 | Rhamnose, fucose, xylose, mannose | 0.47±0.10* | 0.70±0.30 |
| Cesa6 c | At5g64740 | Cellulose | 1.28±0.11* | 3.04±0.29* |
| cesa8 d | At4g18780 | Cellulose | 2.73±0.24* | 3.51±0.32* |
| xxt1/xxt2 e | At4g02500/At3g62720 | Xyloglucan | 4.13±0.36 | 2.43±0.58 |
| Family 77 GTs | ||||
| lrd5-2 | At2g35610 | Extensin arabinose | 0.77±0.11* | 0.80±0.22* |
| rra1 f | At1g75120 | Arabinose | 0.10±0.04 | – |
| rra2 f | At1g75110 | Arabinose | 0.07±0.03 | – |
| ray1 g | At1g70630 | AGP arabinose | 5.21±0.54* | 0.85±0.18* |
| lrd5-2/ray1 | At2g35610/At1g70630 | – | 5.16±0.42* | 2.12±0.28* |
a Reiter et al., 1997; b Austin et al., 2011;c Turner and Somerville, 1997; d Fagard et al., 2000; e Cavalier et al., 2008; f Egelund et al., 2007; g Gille et al., 2013
Fig. 9.
Phenotypes of cell wall mutants grown for 14 d on repressive conditions. Many cell wall mutants show lateral root phenotypes.
To further investigate the relationship between cell wall properties and lateral root formation, a panel of mutants previously characterized to have altered cell wall composition were screened for lateral root phenotypes (Fig. 9). Attention was focused on the murus (mur) series of mutants, as well as prc1 (cesa6), irx1 (cesa8), and xxt1/xxt2 (Reiter et al., 1997; Turner and Somerville, 1997; Fagard et al., 2000; Cavalier et al., 2008). This set includes mutants with altered levels of cellulose, xyloglucan, fucose, arabinose, rhamnose, xylose, and/or mannose in the cell wall (all but one of the mutants altered in arabinose have not been cloned and therefore it is unknown if they are altered in the same pathway as lrd5). Interestingly, nine out of 14 mutants in this set showed an increase in lateral root formation compared to the wild-type background (Col) when grown under repressive conditions (Table 1). Lateral root formation was also analyzed in each of the cell wall mutants grown on parafilm to restrict contact between the leaves and the nutrient media, to reveal whether differences in shoot uptake of media sucrose could account for the root system phenotypes, as has been seen previously for mutants in cuticle formation (Macgregor et al., 2008). While the phenotypes of mur3, mur9, and mur 11 might be attributable to this phenomenon, the remaining mutants tested still showed increased lateral root formation when their shoots were isolated on parafilm, indicating that these mutants form increased numbers of lateral roots independent of nutrient uptake through aerial tissues. Together, these results demonstrate that the phenotype seen in lrd5/xeg113 is not specific to cell wall extensin arabinosylation or to cell wall arabinose levels, but is shared with many mutants that compromise cell wall biosynthesis.
Discussion
A pipeline for categorizing mutations with direct and indirect effects on lateral root formation
Lateral root formation can be broadly considered to involve lateral root initiation, primordia development and patterning, and primordia emergence. Mutations in any of these processes (and others) manifest as altered numbers of lateral roots. This study demonstrates a pipeline that distinguishes between the mechanisms compromised in a set of eight mutants with increased lateral root formation. Such a pipeline should be broadly useful for understanding whether lateral root mutant phenotypes are an indirect result of altered plant/shoot system growth, are a result of shoot tissues that are ‘leaky’ to sucrose in culture media, or can truly be described as identifying genes involved in root development. It allowed the identification of lrd5/xeg113 as a mutant specifically affected in lateral root development and emergence.
The role of LRD5/XEG113 in constraining lateral primordium emergence
Gille et al. (2009) demonstrated that LRD5/XEG113 is involved in elongation of arabinoside chains on extensins. Extensins are hydroxyproline-rich glycoproteins that are embedded in the plant cell wall, and it is believed that their role is strictly to provide mechanical reinforcement (Lamport, 2001). This suggests that the effect of LRD5/XEG113 on lateral root formation is mediated by its role in the physical characteristics of the cell walls. Given that extensins are thought to strengthen the cell wall, one possibility is that cell wall modifications catalysed by LRD5/XEG113 make it more difficult for primordia to push between overlying cell layers during emergence. More recent work has shown that XEG113 and other enzymes involved in extensin modification play an important role in polarized cell growth in root hairs and pollen tubes (Velasquez et al., 2011; Ogawa-Ohnishi et al., 2013). However, the idea that LRD5/XEG113 contributes to a broad characteristic of the cell wall, such as strength, is more consistent with the fact that many unrelated mutants in cell wall synthesis demonstrate similar lateral root phenotypes. This makes it unlikely that the lrd5 phenotype is specifically associated with modification of extensins or arabinose chain length.
Transgenic plants expressing LRD5/XEG113:GFP fusions under the control of an endogenous promoter suggest that, in the root, LRD5/XEG113 is expressed most strongly in trichoblasts near the root tip. This has not been previously demonstrated, but is strongly predicted by microarray experiments (http://www.arexdb.org). Furthermore, trichoblast expression is consistent with the demonstrated important role of XEG113 in root hair growth (Velasquez et al., 2011). Therefore, the reporter lines are likely to accurately reflect LRD5/XEG113 expression. It seems unlikely that the loss of LRD5/XEG113 from trichoblasts is responsible for increased lateral root primordia emergence in the lrd5 mutants. Instead, it is hypothesized that low levels of expression in other cell types at the root tip, also predicted by microarray, are responsible. In this model, critical cell wall characteristics are established in young cells that will come to overlay lateral root primordia later in development. Rescue of the lrd5 phenotype with tissue-specific expression will be necessary to test this model. However, the preferential expression of LRD5/XEG113 in young trichoblast cells, long before root hair formation begins, suggests that the early events in cell wall formation can be manifested later in development.
The role of the cell wall in constraining lateral primordium emergence
Some cell wall mutants affected lateral root primordia emergence while others did not. The set that showed a lateral root phenotype included prc1 and ixr1 (altered in cellulose deposition), xxt1/xxt2 (no detectible xyloglucan), mur1 (reduced fucose in xyloglucan, rhamnogalacturonan II, and arabinogalactan proteins), mur3 (reduced fucose in xyloglucan), mur4 (almost no arabinose in rhamnogalacturonan I and II, glucoarabinoxylans, hydroxyproline-rich proteins, and arabinogalactan proteins), and mur8, mur9, and mur11 (three mutants altered in cell wall rhamnose, fucose, xylose, and mannose content) (Bonin et al., 1997; Reiter et al., 1997; Turner and Somerville, 1997; Fagard et al., 2000; Vanzin et al., 2002; Burget et al., 2003; Bosca et al., 2006; Cavalier et al., 2008). Only two of these mutants had been previously reported to have lateral root phenotypes (van Hengel and Roberts, 2002; Hermans, 2010). This data raises the question of what these mutants have in common that distinguishes them from cell wall mutants that fail to show a lateral root phenotype. It is possible that genes affecting lateral root formation act to strengthen the walls of cells overlying primordia, as hypothesized here for LRD5/XEG113. All of the genes perturbed in the tested mutants that have altered lateral root emergence (and have been cloned) are predicted to be expressed in cells that at some point overly primordia (www.arexdb.org). mur10, on the contrary, is not expressed in cells overlying primordia and does not have a lateral root phenotype. Although not conclusive, this observation is consistent with the hypothesis that alterations in the cells that overly primordia are responsible for the emergence phenotype seen in the mutants. Mechanical tests on hypocotyls in mur1, mur3, mur10, and xxt1/xxt2 have shown that these mutants have altered cell wall strength (Ryden et al., 2003; Pena et al., 2004; Bosca et al., 2006; Cavalier et al., 2008; Abasolo et al., 2009). In contrast, mur2, although altered in the fucosylation of xyloglucan along with mur1 and mur3, is only marginally weaker in mechanical tests compared to the wild type (Ryden et al., 2003; Pena et al., 2004; Abasolo et al., 2009) and does not have a lateral root phenotype. This may imply that the reduction in strength in mur2 is insufficient to cause an alteration in lateral root emergence. Despite these correlations, it remains possible that a broad change in cell wall composition is shared by mutants with lateral root phenotypes and that this change affects lateral root formation by a mechanism independent of cell wall strength. Indeed, in the lrd5 mutant, all stages of lateral root development were accelerated (Fig. 6). This might be evidence that cell walls play a role in growth and development that is more complex than creating physical constraint.
The notion that the walls of cells overlying primordia are important in restricting emergence has been proposed previously in the literature (Neuteboom et al., 1999; Laskowski et al., 2006; Gonzalez-Carranza et al., 2007; Swarup et al., 2008; Peret et al., 2009). A similar role was recently shown for the root casparian strip, which apparently constrains the transition of the lateral root primordia from a flat to rounded morphology during development (Lucas et al., 2013). However, the present work is the first, as far as is known, to suggest that a weakening or modification of the cell wall is sufficient to promote lateral root primordia emergence. This would mean that lateral root primordia are poised for emergence and are constrained merely by the presence of overlying layers of cell walls. This is consistent with the model of Peret et al. (2009), who proposed that, during normal development, LAX3 expression in cells overlying the primordia allows auxin to induce cell wall remodelling enzymes in these cells, leading to cell separation and lateral root primordia emergence. This is also consistent with the observations of Lucas et al. (2013), who showed that expression of a dominant negative AXR3 allele in tissues overlying a primordium delayed development, which they interpret to mean that primordia cannot emerge in the absence of auxin-induced cell wall remodelling.
This study suggests that mutations, environmental response pathways, or intrinsic pathways that weaken or otherwise alter cell walls may be sufficient to promote lateral root formation by facilitating the separation of the cells overlying the lateral root primordium. One way to test this model further would be to introduce the lrd5/xeg113 or ray1 mutations into a lax3 aux1 double-mutant background to see if cell wall defects in lrd5 or ray1 could compensate for the putative loss of a mechanism to soften cell walls. However, triple mutant progeny could not be recovered from either aux1/aux1 lax3/lax3 lrd5/+ or ray1/+ lines. This suggests a strong interaction between auxin import and cell wall composition that will require further work to decipher.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Shoot systems of eight mutants isolated based on increased lateral root formation.
Supplementary Fig. S2. Correlation of plant shoot mass and absorbance of ethanol extracts.
Supplementary Fig. S3. Lateral root emergence in a second lrd5 allele and after rescue.
Acknowledgements
The authors thank Marcus Pauly, Sascha Gille, Paul Ingram, Steve Bernacki, Anjoli Doshi, and Namrata Garg for their contributions to this work. This paper is dedicated to the friends and family of Peter Roycewicz.
References
- Abasolo W, Eder M, Yamauchi K, et al. 2009. Pectin may hinder the unfolding of xyloglucan chains during cell deformation: implications of the mechanical performance of Arabidopsis hypocotyls with pectin alterations. Molecular Plant 2, 990–999 [DOI] [PubMed] [Google Scholar]
- Austin RS, Vidaurre A, Stamatiou G, et al. 2011. Next-generation mapping of Arabidopsis genes. The Plant Journal 67, 715–717 [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD. 1997. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proceedings of the National Academy of Sciences, USA 94, 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. 2006. Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiology 142, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget EG, Verma R, Molhoj M, Reiter WD. 2003. The biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis . The Plant Cell 15, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. 2003. Dissecting Arabidopsis lateral root development. Trends in Plant Science 4, 165–171 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, et al. 2008. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. The Plant Cell 20, 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta . Plant Physiology 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I. 2012. Lateral root initiation: one step at a time. New Phytologist 4, 867–873 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inzé D, Beeckman T. 2006. Lateral root initiation or the birth of a new meristem. FEBS Letters 6, 871–887 [DOI] [PubMed] [Google Scholar]
- Deak KI, Malamy J. 2005. Osmotic regulation of root system architecture. The Plant Journal 43, 17–28 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P. 2000. Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana . Planta 214, 30–36 [DOI] [PubMed] [Google Scholar]
- Egelund J, Damager I, Faber K, Olsen CE, Ulvskov P, Petersen BL. 2008. Functional characterisation of a putative rhamnogalacturonan II specific xylosyltransferase. FEBS Letters 582, 3217–3222 [DOI] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL. 2007. Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. FEBS Letters 64, 439–451 [DOI] [PubMed] [Google Scholar]
- Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N. 2006. Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-alpha-d-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. The Plant Cell 18, 2593–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H. 2000. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis . The Plant Cell 12, 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA. 1991. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. The Plant Journal 1, 71–82 [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis . The Plant Journal 44, 382–395 [DOI] [PubMed] [Google Scholar]
- Gille S, Hansel U, Ziemann M, Pauly M. 2009. Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proceedings of the National Academy of Sciences, USA 106, 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Sharma V, Baidoo EEK, Keasling JD, Scheller HV, Pauly M. 2013. Arabinosylation of a yariv-precipitable cell wall polymer impacts plant growth as exemplified by the Arabidopsis glycosyltransferase mutant ray1 . Molecular Plant 6, 1369–1372 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Carranza ZH, Elliott KA, Roberts JA. 2007. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana . Journal of Experimental Botany 58, 3719–3730 [DOI] [PubMed] [Google Scholar]
- Hermans C, Porco S, Verbruggen N, Bush DR. 2010. Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiology 152, 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, McCouch SR. 2013. Getting to the roots of it: genetic and hormonal control of root architecture. Frontiers in Plant Science 4, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S, Runions J, Köhler U, Laplaze L, Hodge S, Haseloff J. 2005. Marking cell lineages in living tissues. The Plant Journal 42, 444–453 [DOI] [PubMed] [Google Scholar]
- Lamport DT. 2001. Life behind cell walls: paradigm lost, paradigm regained. Cellular and Molecular Life Sciences 58, 1363–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. 2006. Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant and Cell Physiology 47, 788–792 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463 [DOI] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, et al. 2013. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proceedings of the National Academy of Sciences, USA 110, 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor DR, Deak KI, Ingram PA, Malamy JE. 2008. Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. The Plant Cell 20, 2643–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell and Environment 28, 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE. 2009. Lateral root formation. In: Beeckman T. (ed), Annual plant reviews volume 37: root development. Oxford: Wiley-Blackwell; pp 83–126 [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development 124, 33–44 [DOI] [PubMed] [Google Scholar]
- Neuteboom LW, Veth-Tello LM, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ. 1999. A novel subtilisin-like protease gene from Arabidopsis thaliana is expressed at sites of lateral root emergence. DNA Research 6, 13–19 [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y. 2013. Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana . Nature Chemical Biology 9, 726–730 [DOI] [PubMed] [Google Scholar]
- Pena MJ, Ryden P, Madson M, Smith AC, Carpita NC. 2004. The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiology 134, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14, 399–408 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR. 1997. Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. The Plant Journal 12, 335–345 [DOI] [PubMed] [Google Scholar]
- Roycewicz P, Malamy JE. 2012. Dissecting the effects of nitrate, sucrose and osmotic potential on Arabidopsis root and shoot system growth in laboratory assays. Philosophical Transactions of the Royal Sociiety London B 367, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden P, Sugimoto-Shirasu K, Smith AC, Findlay K, Reiter WD, McCann MC. 2003. Tensile properties of Arabidopsis cell walls depend on both a xyloglucan cross-linked microfibrillar network and rhamnogalacturonan II-borate complexes. Plant Physiology 132, 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, et al. 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nature Cell Biology 10, 946–954 [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. 1997. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. The Plant Cell 9, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. 2002. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis . The Plant Journal 32, 105–113 [DOI] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. 2002. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1 . Proceedings of the National Academy of Sciences, USA 99, 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, et al. 2011. O-Glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Molecular Biology and Evolution 21, 809–818 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.