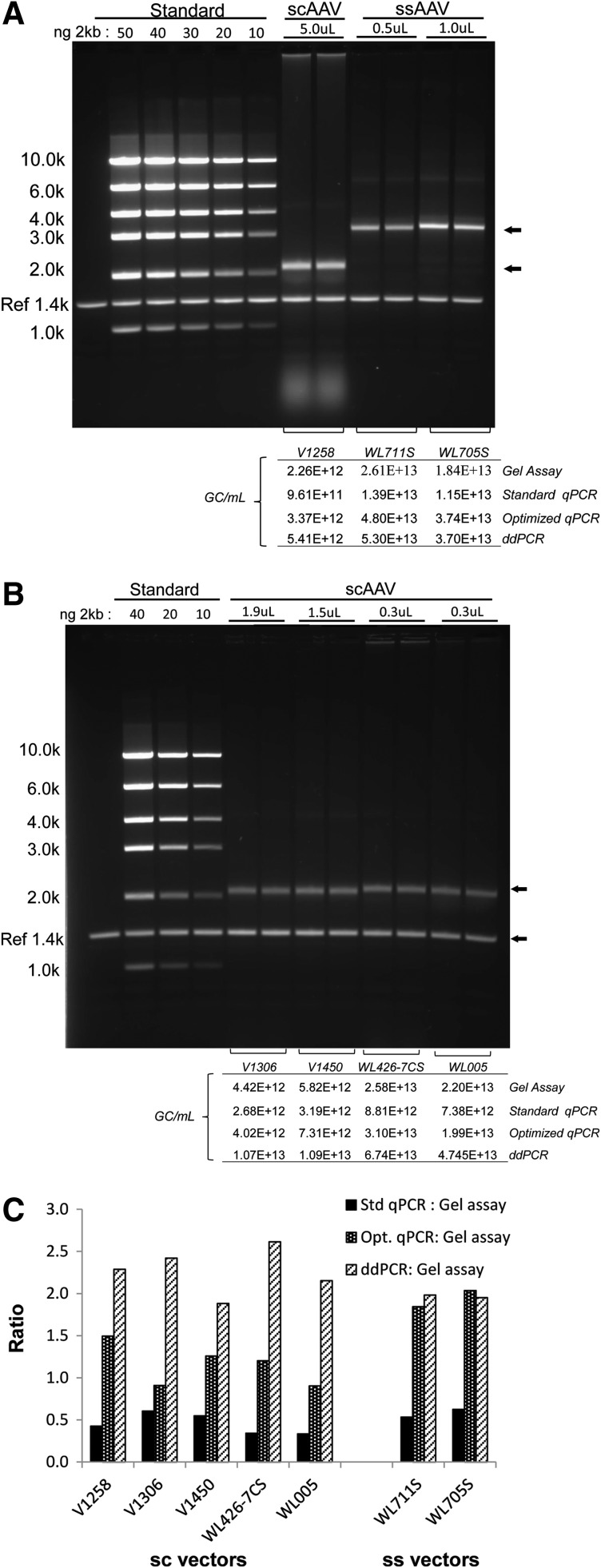
FIG. 3.
Native agarose gel genome titer assay. (A) The assay was performed according to the published method of Fagone and colleaguges (2011) and as described in Materials and Methods. A dilution series of DNA mass markers was run alongside duplicate lanes of one scAAV and two ssAAV vector samples and used to quantify the full-length double-stranded genome band (arrows). An internal reference marker (Ref 1.4k) was run in each lane and used to normalize loading between lanes. The gels were quantified and gel titers were calculated as described in Materials and Methods. The amount (nanograms) of the 2-kb standard fragment and the volume (microliters) of each vector loaded are indicated above the respective lanes. The table below the lanes shows the calculated titer for the vectors analyzed by the gel assay and compares them with titers calculated by each of the PCR-based assays using primer–probe sets targeting the poly(A) signal sequence. (B) Four additional scAAV vectors (duplicate lanes) and standards as described previously were run on native agarose gels. The calculated gel assay titers in comparison with PCR-based assay titers [poly(A) signal sequence] are shown in the table below the lanes. (C) Ratios between titers obtained by PCR-based assays and the native gel assay for scAAV and ssAAV vector lots are shown.