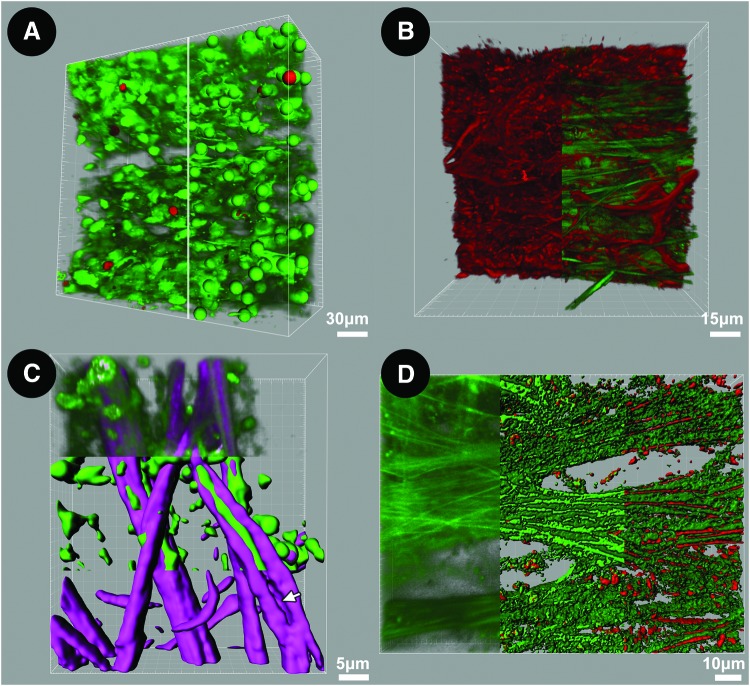
FIG. 11.
Three-dimensional (3D) reconstruction and analysis of human trabecular meshwork (TM). (A) 3D reconstruction and analysis of live/dead co-labeling by calcein AM (green) and propidium iodide (PI; red) in TM. Tangential view (246×246 μm) is shown, with uveal meshwork seen on the upper face. Left half of image: Labeled cells are predominantly alive [calcein-positive (green) and PI-negative labeling (red) is scant]. Right half of image: spot mapping of individual cells during which different spheres are assigned to live (green) and dead (red) cells to allow automated live/dead cell counting. (B) Multimodal imaging combining F-actin epifluorescence (red filaments; left half of image) and structural matrix autofluorescence (AF); green fibers; right half of image). Actin filaments form an interconnected network among autofluorescent trabecular beams. (C) Molecular dissection of structural matrix by multimodal imaging and “isosurface” modeling. Model shows collagen (second harmonic generation (SHG); purple) encasing elastic fibers (AF; green) in the uveal meshwork. Upper third: 3D reconstruction of original SHG and AF signals; middle third: isosurface mapping segregating AF and SHG; lower third: SHG isosurface map after subtraction of AF signal to show space occupied by elastin in beam (arrow). (D) Mapping of proteins corresponding to components of AF signal. Left third: 3D reconstruction of original AF signal; middle third: isosurface model segregating high- and lower-intensity AF signals; right third: co-localization of eosin fluorescence (elastin) with high-intensity AF but not lower-intensity AF. Color images available online at www.liebertpub.com/jop