Summary
Because it appears that almost all organisms are part of an interdependent meta-organism, an understanding of the underlying host-microbe species associations, and of its evolution and molecular underpinnings, has become the new frontier in zoology. The availability of novel highthroughput sequencing methods together with the conceptual understanding that advances mostly originate at the intersection of traditional disciplinary boundaries, enable biologists to dissect the mechanisms that control the interdependent associations of species. In this perspective article, we outline some of the issues in interspecies interactions, present two case studies illuminating the necessity of interfacial research when addressing complex and fundamental zoological problems, and show that an interdisciplinary approach that seeks to understand co-evolved multi-species relationships will connect genomes, phenotypes, ecosystems and the evolutionary forces that have shaped them. We hope that this article inspires other collaborations of a similar nature on the diverse landscape commonly referred to as “zoology”.
Interfacial research to thoroughly understand host-microbe species interactions
In physics, the interface between two phases has very different properties from that of the bulk phase and is important in a variety of processes; many natural and technological processes involve phenomena dominated by interfacial mechanics, that is, occurring within the regions of intersection beween several fluid and /or solid phases. In contemporary zoology, specialization still has its place but the dynamics of interdisciplinary approaches appear central to approaching some of the toughest problems, such as a successful explanation of the biochemical basis of adaptations, the origin of morphological novelty, and a thorough understanding of species interactions, among and between members of the macro- and microbiota.
Technological advances driven by researchers representing multiple disciplines have altered our perspective on zoological questions. The potential uses and applications of next-generation sequencing technologies, for example, span the whole spectrum of ecological and evolutionary research and enable to approach the interplay between genes and environment. For many animal species genomic resources are available including genomic and cDNA libraries, microarrays, web-based bioinformatic portals, annotation and gene expression databases, and well-resolved phylogenetic frameworks. Extensive genomic and transcriptomic analyses of single cells (using whole genome amplification) to entire microbial communities (using metagenomics and metatranscriptomics) are now feasible because of ultrahigh throughput sequencing methods at extremely low costs. Obtaining the complete genomic sequence information of a whole ecosystem may be in reach within the next 10 years. In parallel to these advances in molecular techniques, novel high-resolution imaging methods, such as fluorescence techniques at subcellular resolution, digital-enhanced polarization, frequency and time resolved spectroscopic techniques, allow the quantitative analysis of cellular responses on external factors on the level of single proteins. A rapid expansion in the application of mathematics and analytical methodologies of computer science to biology allows advanced computational modelling and simulation to describe complex interacting systems in mathematical or statistical terms.
Metaorganisms – a new term for an old concept
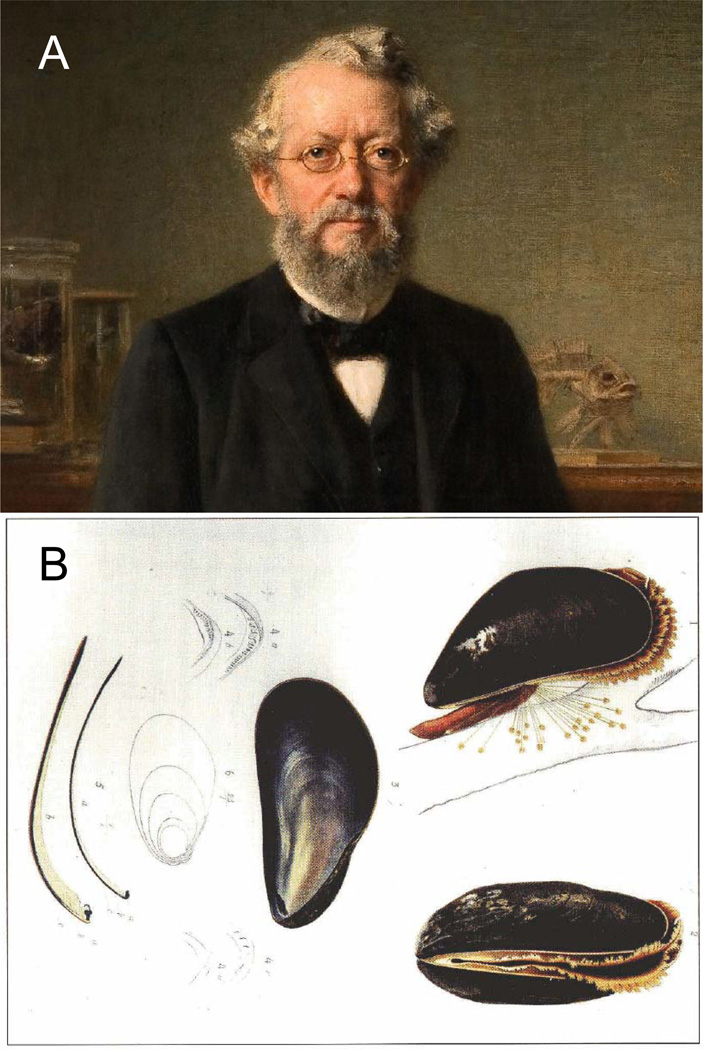
In 1877, Karl Möbius, Professor of Zoology at Kiel University (Figure 1), coined the term “biocenosis” for a community of living beings belonging to different species and associated by way of inter-species interdependence. In one of the first studies, later to become a classic, to be conducted in the emerging science of ecology, Möbius was seeking to determine why the oyster beds of Cancale, Marennes and Arca-chon were becoming exhausted, while the oyster beds in the British river estuaries and the Schleswig-Holstein oyster beds were very rich (Möbius, 1877). He related this phenomenon to the other species present, rather than to the oysters in the beds themselves. Möbius thus was the first to recognise that an ecological system must be taken as a whole and coined the term "biocenosis" for a living community. His biocenosis theory established itself as the basis of general ecology.
Figure 1. Karl August Möbius and the biocenosis concept.
(A) Karl August Möbius, Gemälde von Ernst Hildebrand, 1895. (B) Möbius´ study object: the molluscs in the North Sea. From: H. A. Meyer, K. Möbius: Fauna der Kieler Bucht. Erster Band: Die Hinterkiemer oder Opisthobranchia. Zweiter Band: Die Prosobranchia und Lamellibranchia nebst einem Supplement zu den Ophistobranchia. Engelmann. Leipzig 1865–1872
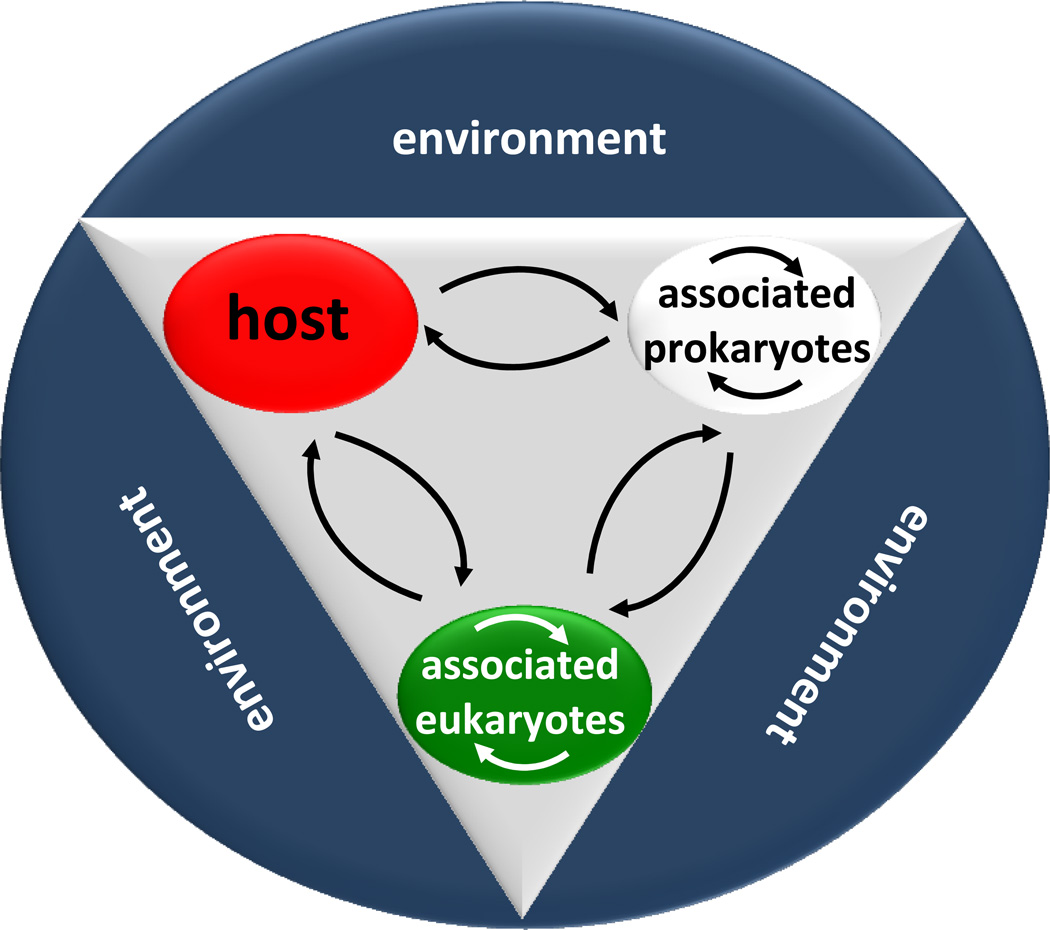
Today we realize that any multicellular organism must be considered a meta-organism comprised of the macroscopic host and synergistic interdependence with bacteria, archaea, fungi, and numerous other microbial and eukaryotic species including algal symbionts (Figure 2). We specifically refer to these associations as "metaorganisms", because this collective term defines a superordinate entity that is applicable to all kinds of interdependent associations and the term is not constrained to specific taxonomic groups such as "holobiont" (usually used for cnidarians; for references see for example: Rohwer et al., 2002; Vega Thurber et al., 2009; Bourne et al., 2009) or "superorganism" (used for social insects such as ants; see for example Behmer 2009). Metaorganisms are polygenomic organisms. The term “metaorganism” was first used by Graham Bell (1998) to refer to organisms which are between two levels of organization. Recently, the term is increasingly used to refer to the totality of any multicellular organism derived from millennia of co-evolution with microbiota (Biagi et al., 2011). Even humans have been reviewed as ‘metaorganisms’ as a result of a close symbiotic relationship with the intestinal microbiota (Turnbaugh et al., 2007).
Figure 2.
Multicellular organisms are metaorganism comprised of the macroscopic host and synergistic interdependence with bacteria, archaea, fungi, and numerous other microbial and eukaryotic species including algal symbionts.
Microbes as elements of a forgotten organ
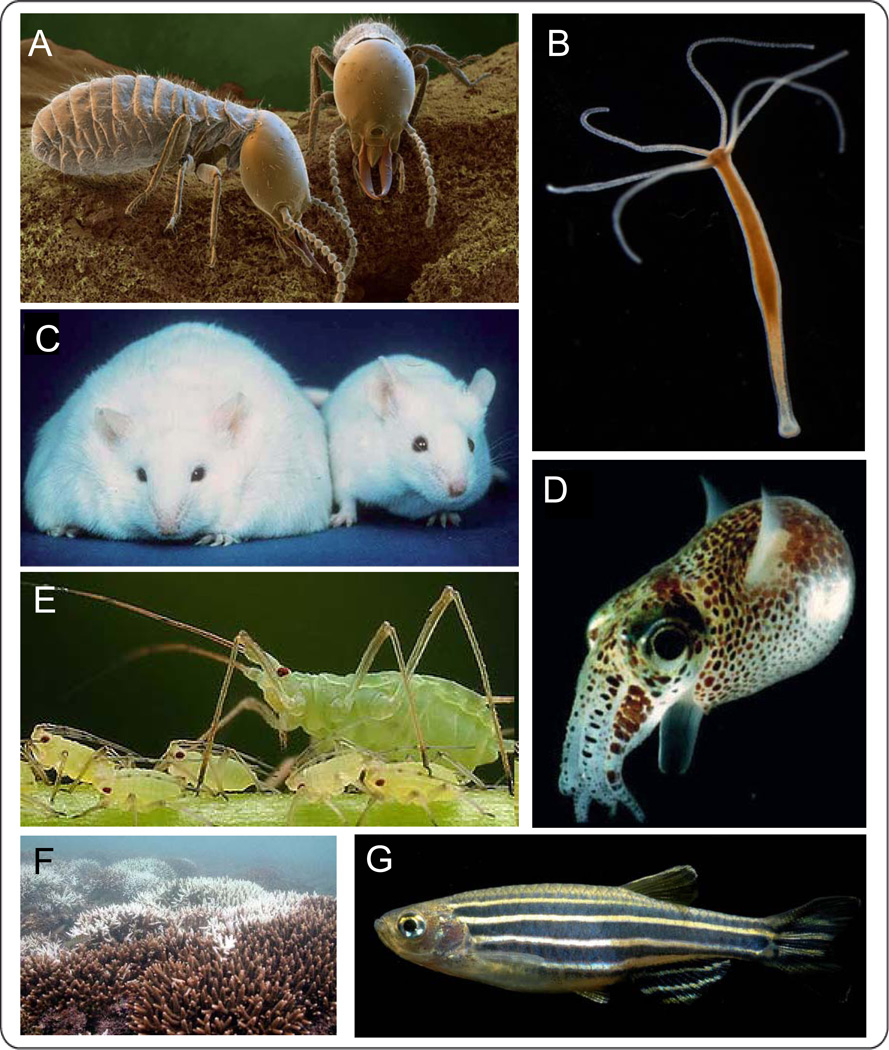
From protists to humans, all animals and plants are inhabited by microbial organisms. Most of life´s diversity in fact originates from microbial organisms. It is therefore remarkable that the microbial world is so little recognized by the zoologist except in the context of animal or plant disease and environmental decomposition. The reason for the general neglect is obvious: perception of the microbial world usually requires observation of the consequences of microbial activities, which can be subtle and difficult to investigate experimentally (McFall-Ngai, 2002; 2008). However, there is an increasing appreciation that microbes are an essential part of the animal phenotype influencing fitness and thus ecologically-important traits of their hosts (O’Hara AM and Shanahan F, 2006; McFall-Ngai, 2007; Fraune and Bosch, 2010). Macroscopic manifestations of microbial activities in animal development can be found in an increasing number of invertebrate and vertebrate models (Figure 3). Now, with an array of high-resolution technologies, first insights into the crosstalk between animal hosts and their microbial symbionts for maintaining a long-term coexistence have been reported (Wier et al., 2010).
Figure 3. Selected model organisms allow integrated analysis of host-symbiont interactions.
(A) Termites live in symbiosis with cellulose-degrading gut microbes (© Bayer). (B) Hydra is a new model organism for epithelial host-microbe interactions. (C) Pathogenic effects of a disturbed host-microbe homeostasis have been discovered in mice (© ONRL). (D) Aphids harbor the symbiont Buchnera aphidicola, which supplies the host with essential amino acids, that are not included in the phloem sap diet (© Alex Wild). (E) The bacterium Vibrio fisherii induces the formation of the light organ in the squid E. scolopes (picture taken from M. J. McFall-Ngai and E. G. Ruby). (F) The coral bleaching disease is a global threat of coral reefs. Changes in the microbiota of the coral host can confer resistance to bleaching (© Lee James Pantas). (G) The gut development of the zebrafish Danio rerio is dependent on the presence of gut bacteria.
Many animals carry symbiotic microbes that provide defense against natural enemies (Gill-Turnes et al., 1989; Arnold et al., 2003; Jaenike et al., 2010). In Drosophila, for example, the endosymbiont Spiroplasma rescues D. neotestacea females from the sterilizing effects of nematode parasitism. In vertebrates, Chytridiomycosis, a fungal disease caused by Batrachochytrium dendrobatidis (Bd), has caused many amphibian declines and extinctions. Resistance in amphibians unaffected by Bd is in part due to the presence of antifungal microbial species on the skin. Lisa Belden´s lab at Virginia Tech in Blacksburg has recently isolated the bacterial species Janthinobacterium lividum from three temperate amphibian species including Rana musoca and shown in vitro that it inhibits Bd by secreting violacein, an antimicrobial metabolite, at low concentrations (Jenifer Walke, pers. commun.). Members of the normal human microbiota also inhibit pathogen colonization in human: Staphylococcus epidermidis, for example, inhibits nasal colonization by Staphylococcus aureus, a human opportunistic pathogen (Iwase et al., 2010) via a serine protease. From an evolutionary perspective, these examples demonstrate that in natural populations defensive symbionts must be considered as major players in the ecology of species interactions (Gil-Turnes et al 1989; Arnold et al., 2003; Oliver et al., 2003; Scarborough et al., 2005; Hedges et al., 2008; Teixeira et al., 2008; Iwase et al., 2010).
Bacteria also must be seen as essential part of the vertebrate immune system. The paradigm that the adaptive immune system has evolved to control microbes has been modified to include the concept that the immune system is in fact controlled by microorganisms (McFall-Ngai, 2007; Eberl, 2010). For instance, access for microbiota to the adaptive immune system in mammals has beneficial consequences for the host, in that antibacterial TH17 and TH1 responses are generated that are known to control pathogenic bacterial pathogens (Ivanov et al., 2008; Ivanov et al., 2009). The induction of TH17 and TH1 responses in the gut is closely associated with the induction of regulatory T cell subsets (Chaudhry et al., 2009). These responses are known to have beneficial effects for survival of long-lived chronic helminth infections in the absence of overt pathology and indeed are central to the tenet of the modified hygiene hypothesis (Yazdanbakhsh & Matricardi, 2004).
The diverse interactions reported between the mammalian gut and its microbiota include not only the control of the immune system but also cooperation in food breakdown (e.g. Hooper et al. 2002), participation in the regulation of fat accumulation (Bäckhed et al. 2004), and the induction of specific gene expression in the intestinal cells (e.g., Cash et al.,2006); microbes are also needed for the formation of the villi capillaries (Stappenbeck et al. 2002) and gut-associated lymphoid tissue (Bouskra et al., 2008).
Species interactions moderate evolutionary processes
A strictly microbe-dependent life style has profound evolutionary consequences – and implies that the phenotype of a healthy animal cannot be explained entirely by its genome. Based on field and laboratory observations indicating that corals can adapt rapidly to changing environmental conditions by altering their population of symbiotic bacteria (Kushmaro et al., 1996; Koren and Rosenberg, 2006; Rosenberg et al., 2007), a group of scientists at Tel Aviv University proposed (Reshef et al. 2006) that a dynamic relationship exists between symbiotic microorganisms and environmental conditions that brings about the selection of the most advantageous coral holobiont. The hologenome theory of evolution (Zilber-Rosenberg and Rosenberg, 2008) considers the holobiont with its hologenome as a unit of selection in evolution. While the holobiont is defined as the host organism and all of its symbiotic microbiota, the hologenome is the sum of the genetic information of the host and its microbiota. The theory is based on the facts that all animals and plants harbour abundant and diverse microorganisms; that symbiotic microorganisms are transmitted between generations; and that under environmental stress, the symbiotic microbial community can change rapidly. The hologenome theory suggests that microbial symbionts affect the fitness of the holobiont and play an important role both in adaptation and in evolution of higher organisms. Genetic variation in the holobiont that can occur either in the host and/or in the microbial symbiont genomes can then be transmitted to offspring. In addition to the known modes of variation, i.e., sexual recombination, chromosomal rearrangement and mutation, variation in the holobiont can occur also via two mechanisms that are specific to the hologenome theory: amplification of existing microorganisms and acquisition of novel strains from the environment (Rosenberg et al., 2009). Thus, rapid changes in the symbiotic microbiota could allow the holobiont to adapt and survive under changing environmental conditions. But why do multicellular organisms have such different microbial communities? And how much do they matter? Do different organisms have distinct microbial signatures at birth, or do they evolve as the organisms age? While these questions remain to be anwered, integrative and interdisciplinary approaches in two invertebrate metaorganisms, corals and aphids, have uncovered surprising links between host-microbe interactions and defense strategies.
Case study 1: Species interactions in corals
Coral tissue and coral mucus contains abundant and highly complex microbial communities (Dinsdale et al., 2008 a; Dinsdale et al., 2008b; Kvennefors et al., 2010; Sharp et al., 2010). Warming waters are triggering coral bleaching and disease in the Caribbean, Indian Ocean and Great Barrier Reef off the Australian coast. Coral bleaching is accompanied by drastic shifts in the microbial community factors including changes in the production of antimicrobial compounds (Kushmaro et al., 1996; Frias-Lopez et al., 2002; Rohwer et al., 2002; Ritchie, 2006; Bourne et al 2008; Rosenberg et al., 2009). However, up to now there is little if any understanding about the the coral-microbe interactions and the underlying mechanism for what causes coral disease and bleaching (Ritchie, 2006; Ainsworth et al., 2008; Kvennefors et al., 2008).
It needed an interdisciplinary lab headed by an evolutionary ecologist and research combining mathematical simulations and collaboration with experimental biologists to identify the mechanisms that drive the dynamics of species interactions in the coral reef ecological system (Mao-Jones et al., 2010). The researchers from Cornell University used models to simulate bacterial community dynamics within the surface coral mucus under normal conditions and under warmer conditions. The model reveals for the first time how a healthy, normal microbial community in the coral surface-mucus layer protects corals from disease by preventing the invasion and overgrowth of pathogenic bacteria. When corals are stressed by warmer temperatures, there is a critical threshold where the community of microbes suddenly switches. Species associated with a healthy coral organism -- "resident species" -- decline as pathogens associated with coral disease take their place. The new model also added support to the previous observation that once the disease-causing microbes establish themselves, they persist even if the water cools down enough to favor the beneficial bacteria. The coral is then often too damaged to recover, and the reefs begin to die. In sum, by uniting researchers from multiple disciplines for the first time, there is an explanation how beneficial bacteria on coral suddenly give way to pathogens when waters warm.
Case study 2: Species interactions in aphids
Pea aphids (Figure 3 E) and their symbiotic microorganism Buchnera aphidicola have been co-evolving for about 150 MY. Aphids feed on plant juices that they obtain from the phloem tissue of leaves and stems using long piercing mouthparts. Phloem is rich in carbohydrates, but low in the nitrogenous compounds that complex organisms need to make proteins to survive. Symbiotic Buchnera bacteria living inside the aphid provide these missing proteins. In addition to providing essential amino acids that are scarce in its diet, coupled purine metabolism of aphid and Buchnera contributes to the dependence of the pea aphid on this symbiosis (Ramsey et al., 2010). This symbiosis is uniquely amenable to global analyses of interactions between animals and their resident microbiota since it is the only animal symbiosis for which the genomes of both the host and its bacterial symbiotic partners have been sequenced (International Aphid Genomics Consortium (2010); http://www.iagc.org). “Metaorganismal genomics” now opens new roads to understanding the adaptive processes of these animals to adverse conditions.
During the millions of years that these two symbiotic partners have evolved, both the aphid genome and the Buchnera genome has undergone major genomic changes as a result of adapting to intracellular life (reviewed in Brinza et al., 2009). New research suggests (Nikoh et al. 2010) that the aphids' ability to host Buchnera depends on genes they acquired from yet another species of bacteria via lateral gene transfer (LGT). Most transferred genes were closely related to genes from relatives of Wolbachia (Alphaproteobacteria) indicating that aphids utilize a set of duplicated genes acquired from other bacteria in the context of the Buchnera-aphid mutualism. Taken together, these findings impressively demonstrate the interweaving of organisms and their genomes over time and their merging in different ways.
One of the most unexpected findings of the aphid genome sequencing project was the absence of many genes involved in defending the insect from pathogens, parasites and predators. This was surprising as pea aphids are attacked by a variety of natural enemies ranging from fungal pathogens to parasitoid wasps. The aphids vary in their resistance to the wasps, which scientists previously had chalked up to genetic differences between aphids. Unexpectedly, however, Nancy Moran and colleagues in the University of Arizona’s department of entomology (now at the Department of Ecology and Evolutionary Biology at Yale University) have shown that the wasp-resistant aphids owe their lives not to a specific genetic predisposition but to facultative symbiotic bacteria Hamiltonella defense carried inside them (Oliver et al., 2003; Oliver et al., 2005; Oliver et al., 2009; for review see Oliver et al 2010). Such a newly acquired resistance is heritable, because the bacteria get passed down from mother to her offspring (Moran et al., 2005). Consistent with the holobiont theory of evolution (Zilber-Rosenberg et al., 2008) this implies that aphids acquire resistance to natural enemies by picking up bacterial symbionts, rather than having changes in the aphids' genes. A truly transdisciplinary analysis performed by experts from a Department of Entomology, a Department of Biology, two laboratories of Magnetic Resonance and Atomic and Molecular Physics, an Institute of Physics and Mathematics, a School of Biology and Environmental Science, a Department of Biomolecular Medicine, and a Department of Surgery and Cancer, deepened our understanding of the interaction between aphids and B. aphidicola by uncovering the central role of amino acid metabolism in the aphid – Buchnera symbiosis (Wang et al., 2010).
A call for transdisciplinary collaboration in zoology
Organisms do not live in isolation, but have evolved, and continue to evolve, in the context of complex communities and specific environmental conditions. The two case studies presented here show that concerted multidisciplinary efforts are required in the future to examine the relationship between multiple phenotypes and the environmental context of organisms. Evolutionary biologists are increasingly able to integrate information across many organisms, from multiple levels of organization and about entire systems to gain a new integrated understanding that incorporates more and more of the complexity that characterizes interdependent species associations. We are at the beginning of what one may call a fundamental shift in zoological research. As zoology becomes more interdisciplinary in its practice and interdisciplinary groups are increasingly the order of the day, with advances being made at the intersection of traditional discipline boundaries, the metaorganismal structure of animals should provide a promising searching ground, for some principles of metaorganismal organization must transcend the particular organism that happens to occupy a given role at any moment. We predict that within the next 5 years genomics and ecology will get closely connected. And that only when we begin to understand the molecular base for adaptation and interactions of Lebensgemeinschaften (communities of life, Karl Möbius) can we start to comprehend how ecosystems are functioning. Currently we do not yet know the rules of composition of meta-organisms. We do not even know if rules exist in the usual sense. We take the examples presented as auspicious beginnings that trace evolution´s complex fingerprints by pioneering new kind of collaborative research.
Acknowledgements
We apologize to those authors whose work we were unable to cite due to space restrictions. We thank René Augustin for comments on the manuscript and him and Sören Franzenburg for providing figures. Supported by grants from the Deutsche Forschungsgemeinschaft (DFG) and grants from the DFG Cluster of Excellence programs “The Future Ocean” and “Inflammation at Interfaces" (to TCGB), NIH RO1-AI50661 (to MMN), NSF IOS 0817232 (to MMN), WM Keck Foundation (to MMN) and fellowship from the the John Simon Guggenheim Foundation (to MMN) .
References
- Ainsworth TD, Fine M, Roff G, Hoegh-Guldberg O. Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica. ISME J. 2008;2:67–73. doi: 10.1038/ismej.2007.88. [DOI] [PubMed] [Google Scholar]
- Arnold AE, et al. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15649. doi: 10.1073/pnas.2533483100. (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmer ST. Animal behaviour: feeding the superorganism. Curr Biol. 2009;19(9):R366–R368. doi: 10.1016/j.cub.2009.03.033. . Review. [DOI] [PubMed] [Google Scholar]
- Bell G. Model Metaorganism. A book review. Science. 1998;282(5387):248. [Google Scholar]
- Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Ageing of the human metaorganism: the microbial counterpart. Age. 2011 doi: 10.1007/s11357-011-9217-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2(4):350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne DG, Garren M, Work TM, Rosenberg E, Smith GW, Harvell CD. Microbial disease and the coral holobiont. Trends Microbiol. 2009;17(12):554–562. doi: 10.1016/j.tim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456(7221):507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Brinza L, Viñuelas J, Cottret L, Calevro F, Rahbé Y, Febvay G, Duport G, Colella S, Rabatel A, Gautier C, Fayard JM, Sagot MF, Charles H. Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. Comptes Rendus Biologies. 2009;Volume 332(Issue 11):1034–1049. doi: 10.1016/j.crvi.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, McDaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Brito BR, Ruan Y, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA, Rohwer F. Functional metagenomic profiling of nine biomes. Nature. 2008a;452(7187):629–632. doi: 10.1038/nature06810. Erratum in: Nature. 2008 Oct 9;455(7214):830. [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Thurber RV, Willis BL, Azam F, Knowlton N, Rohwer F. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS ONE. 2008b Feb 27;3(2):e1584. doi: 10.1371/journal.pone.0001584. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 2010;3(5):450–460. doi: 10.1038/mi.2010.20. [DOI] [PubMed] [Google Scholar]
- Fraune S, Bosch TCG. Why bacteria matter in animal development and evolution. Bioessays. 2010;32:571–580. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microbiol. 2002;68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Turnes MS, Hay ME, Fenical W. Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science. 1989;246:116. doi: 10.1126/science.2781297. [DOI] [PubMed] [Google Scholar]
- Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [Abstract/Free Full Text] [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host– microbial interactions shape the nutrient environment of the mammalian intestine. Ann Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010 Feb 23;8(2):e1000313. doi: 10.1371/journal.pbio.1000313. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329(5988):212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- Koren O, Rosenberg E. Bacteria associated with mucus and tissues of the coral Oculina patagonica in Summer and Winter. Appl Environ Microbiol. 2006;72:5254–5259. doi: 10.1128/AEM.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Rosenberg E. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol. 2006;72(8):5254–5259. doi: 10.1128/AEM.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A, Loya Y, Fine M, Rosenberg E. Bacterial infection and coral bleaching. Nature. 1996;380:396. [Google Scholar]
- Kvennefors EC, Leggat W, Hoegh-Guldberg O, Degnan BM, Barnes AC. An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev Comp Immunol. 2008;32(12):1582–1592. doi: 10.1016/j.dci.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Kvennefors EC, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O. Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site- and speciesspecificity of common bacterial associates. PLoS One. 2010;5(4):e10401. doi: 10.1371/journal.pone.0010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HA, Möbius K. Erster Band: Die Hinterkiemer oder Opisthobranchia. Zweiter Band: Die Prosobranchia und Lamellibranchia nebst einem Supplement zu den Ophistobranchia. Engelmann. Leipzig. 1865. Fauna der Kieler Bucht; pp. 1865–1872. [Google Scholar]
- Mao-Jones J, Ritchie KB, Jones LE, Ellner SP. How microbial community composition regulates coral disease development. PLoS Biol. 2010;8(3):e1000345. doi: 10.1371/journal.pbio.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Develop Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445(7124):153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. Are biologists in 'future shock'? Symbiosis integrates biology across domains. Nat Rev Microbiol. 2008 Oct;6(10):789–792. doi: 10.1038/nrmicro1982. 2008. [DOI] [PubMed] [Google Scholar]
- Möbius KA. Die Auster und dieAusternwirthschaft. Berlin: Wiegandt, Hempel & Parey; 1877. 1877. [Google Scholar]
- Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci U S A. 2005;102(47):16919–16926. doi: 10.1073/pnas.0507029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, McCutcheon JP, Kudo T, Miyagishima SY, Moran NA, Nakabachi A. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010 Feb 26;6(2):e1000827. doi: 10.1371/journal.pgen.1000827. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1803. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Moran NA, Hunter MS. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci U S A. 2005;102(36):12795–12800. doi: 10.1073/pnas.0506131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325(5943):992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. 2010 Review. [DOI] [PubMed] [Google Scholar]
- Ramsey JS, MacDonald SJ, Jander G, Nakabachi A, Thomas GH, Douglas AE. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010 Mar;19(Suppl 2):241–248. doi: 10.1111/j.1365-2583.2009.00945.x. 2010. [DOI] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The Coral Probiotic Hypothesis. Environ Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucusassociated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coralassociated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nature Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Sharon G, Zilber-Rosenberg I. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol. 2009;11(12):2959–2962. doi: 10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- Scarborough CL, Ferrari J, Godfray HCJ. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [Abstract/Free Full Text] [DOI] [PubMed] [Google Scholar]
- Sharp KH, Ritchie KB, Schupp PJ, Ritson-Williams R, Paul VJ. Bacterial acquisition in juveniles of several broadcast spawning coral species. PLoS One. 2010 May 28;5(5):e10898. doi: 10.1371/journal.pone.0010898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber R, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;211(8):2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Carolan JC, Hao F, Nicholson JK, Wilkinson TL, Douglas AE. Integrated metabonomic-proteomic analysis of an insect-bacterial symbiotic system. J Proteome Res. 2010 Mar 5;9(3):1257–1267. doi: 10.1021/pr9007392. 2010. [DOI] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo Mde F, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A. 2010;107(5):2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanbakhsh M, Matricardi PM. Parasites and the hygiene hypothesis: regulating the immune system? Clin Rev Allergy Immunol. 2004;26(1):15–24. doi: 10.1385/CRIAI:26:1:15. Review. [DOI] [PubMed] [Google Scholar]
- Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Lett. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]