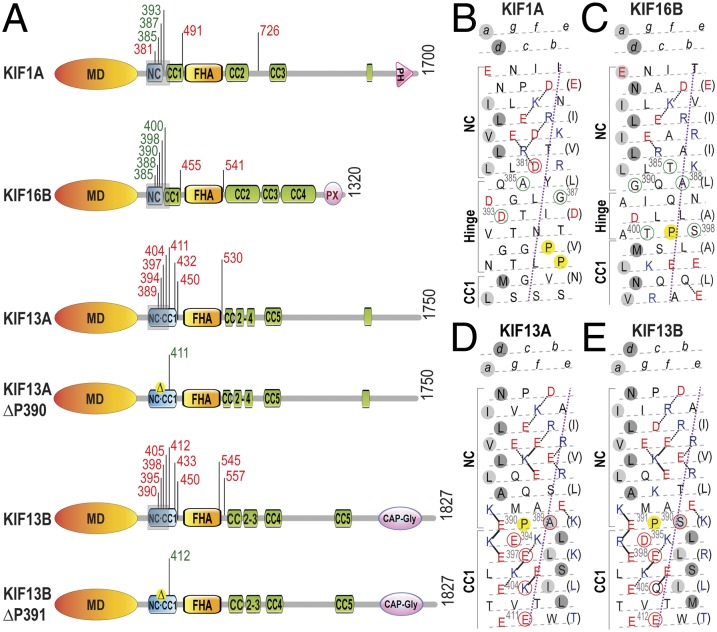
Fig. 2.
The NC is a key regulatory element for kinesin-3 motor dimerization and processive motility. (A) Domain organization of kinesin-3 motors analyzed in this study. Truncations are indicated by black tick marks with the amino acid number. Truncations in green text indicate processive motors, whereas truncations in red text indicate nonprocessive motors based on analysis of motor behavior in CAD cells (Fig. S6) and/or single molecule assays (Fig. 3). CC, coiled-coil; FHA, forkhead-associated; MD, motor domain; NC, neck coil; PH, pleckstrin homology; and PX, Phox homology. (B–E) Heptad net plots of the NC–CC1 regions (gray shaded rectangles in A) of the kinesin-3 motors. The helix has been cut and opened flat to give a 2D representation of the amino acid residues and their potential interactions in the helix. The N terminus is at the top right. Acidic residues are shown in red, basic in blue, and all others in black. Preferred ionic interactions are shown as solid lines and alternative interactions are shown as dotted lines. Every seventh residue is repeated on the right of the plot (in brackets) so that all interactions can be shown. The violet dotted line defines the orientation of the α-helix axis. The light gray circles indicate ‘‘a’’ positions and the dark gray circles indicate ‘‘d’’ positions of the hydrophobic seam in the CC. Residues circled in green indicate truncations that yield processive motors and residues circled in red indicate truncations that yield nonprocessive motors.