Abstract
Malaria is the most important parasitic disease with global concern. Plasmodium knowlesi recently has emerged from its natural simian host as a significant cause of human malaria, particularly in Malaysian Borneo. Therefore, it has been added as the fifth human Plasmodium specie which is widely distributed in Southeast Asia. Recent developments of new molecular tools enhanced our understanding about the key features of this malaria parasite. Here, we review some of the ways in which molecular approaches might be used for epidemiology of P. knowlesi and finally lead to an efficient control of malaria.
Keywords: Malaria, molecular epidemiology, Plasmodium knowlesi
INTRODUCTION
Despite more than a century of efforts to control and eradicate human malaria, it still remains as a major public health problem in the world. The global malaria deaths increased from 995,000 in 1980 to a peak of 1,817,000 in 2004 and declined to 1,238,000 in 2010 because of the scaling up of the control measures especially in Africa.[1] Five species of the genus Plasmodium namely Plasmodium falciparum, Plasmodium vivax, Plasmodium ovale, Plasmodium malariae, Plasmodium knowlesi, are believed to contribute to the human malaria. Zoonotic malaria caused by P. knowlesi has recently been revealed to be a widespread infection in Southeast Asia.[2] To date, over 20 species of Plasmodium are known to be capable of infecting monkeys in which five of them have zoonotic potential including Plasmodium simium, Plasmodium brasilianum, Plasmodium cynomolgi, Plasmodium inui and P. knowlesi.[3] The P. knowlesi-infection has emerged as a common and potentially fatal cause of human malaria in Malaysian Borneo.[2]
The nuclear genome sequence of P. knowlesi is known[4] with close phylogenetic relationship to P. vivax; however, there are important phenotypic differences between them such as length of asexual cycle, host blood cell preference and absence of hypnozoite in P. knowlesi.[5] The 23.5 Mb nuclear genome of P. knowlesi encodes for 5188 proteins which approximately 80% of them has ortholog in P. falciparum and P. vivax.[4] The availability of genomic sequence of several Plasmodium species allows performing comparative genomic analysis and providing insights into the evolution of Plasmodium genes and gene families.[6] Molecular epidemiology is the application of molecular techniques to study pathogen genotypes and gene expression and infer them to the occurrence of infection in human population.[7] Recent new genomic information allows more extensive molecular epidemiological studies for a better control of malaria.
GEOGRAPHICAL DISTRIBUTION OF P. KNOWLESI
P. knowlesi was isolated for the 1st time from a long-tailed macaque monkey (Macaca fascicularis)[8] and the first experimental transmission of this monkey malaria to human was reported by Knowles and Das Gupta in 1932.[9] In 1965, the first natural infection of P. knowlesi in human was reported in an American traveler returning from Peninsular Malaysia.[10] Initially, it was uncertain that the natural human infections could take place and thus P. knowlesi could have zoonotic importance. But later on, experimental mosquito transmission to human and transmission from monkey to human approved the zoonotic potential of P. knowlesi.[11] Until recently, it was believed that natural human cases of P. knowlesi are rare; however, a study by Singh et al. showed that this parasite is a major cause of malaria in Malaysia.[12] Recent molecular studies in several Southeast Asian countries showed the presence of this parasite in human population in Malaysia,[12,13] Thailand,[14] Singapore,[15] Palawan Island in the Philippines,[16] Vietnam,[17] southern Myanmar near the border of China,[18] Indonesian Borneo[19] and Cambodia.[20] There has been an increasing number of imported knowlesi malaria cases in travelers to Southeast Asian countries from the non-endemic countries in Europe,[21,22,23,24,25] USA,[26] New Zealand,[27] Taiwan[28] and Japan.[29]
The long-tailed macaque (M. fascicularis), pig-tailed macaque (Macaca nemestrina) and banded leaf monkeys (Presbytis malalophos) in Southeast Asia are the principal natural hosts of P. knowlesi, although other monkey species are also capable of carrying the parasite.[13] The Anopheles species belong to the leucosphyrus group that have been incriminated as vectors of the parasite, are inhabited in forested areas in Southeast Asia.[30] The geographical range of P. knowlesi corresponds to the overlapping distribution of the vector mosquitoes and the macaque hosts and it defines the risky areas for knowlesi malaria transmission.[13]
APPLICATION OF MOLECULAR METHODS TO DETECT P. KNOWLESI
Conventional microscopy is the primary method for detection and species differentiation of malaria parasites. However, even most skillful microscopists may misdiagnose Plasmodium species in mixed infections especially with morphologically similar malaria parasites.[31] The early trophozoites of P. knowlesi are similar to those of P. falciparum while the mature asexual stages morphologically resemble those of P. malariae underlining the difficulty of identifying knowlesi malaria on the basis of morphology alone and misdiagnosing as P. malariae.[31]
For the first time, P. knowlesi was detected using molecular diagnostic tools, including polymerase chain reaction (PCR) using specific primers for the small subunit ribosomal ribonucleic acid (SSU rRNA), circumsporozoite protein gene and deoxyribonucleic acid (DNA) sequencing [Table 1].[12] Surprisingly, they found that more than half of the P. malariae infection confirmed cases by microscopy were actually P. knowlesi infection and these findings initiated a series of studies in the other Southeast Asian countries to determine the epidemiological distribution of knowlesi malaria.[14,15,16,17,18,19,20] However, Imwong et al. showed that the primers targeting P. knowlesi SSU rRNA cross-react with P. vivax genomic DNA.[32] Therefore, they designed a new set of primers[32] which was used in a semi-nested multiplex PCR identifying the five human Plasmodium species in a three-step reaction.[33] A TaqMan real-time PCR was developed targeting a specific DNA sequence within SSU rRNA of P. knowlesi with the sensitivity of three parasites/μl in the blood and 100% specificity.[34] Moreover, it was proven to be a powerful diagnostic tool for early detection of imported malaria in non-endemic areas.[35] Targeting the same gene, Chew et al. developed a single-step hexaplex PCR system that was able to detect all five human Plasmodium species simultaneously as well as mixed infections up to two-species level.[36] This multiplex PCR was further applied in Sandakan division, Sabah, Malaysia and was able to accurately discover P. knowlesi in the region.[37] In a recent study, a new single-step PCR assay was developed using bioinformatics approach and target multi-copy sequences of P. knowlesi which was able to detect one parasite/μl with 100% specificity.[38]
Table 1.
Summary of molecular methods for specific detection of P. knowlesi
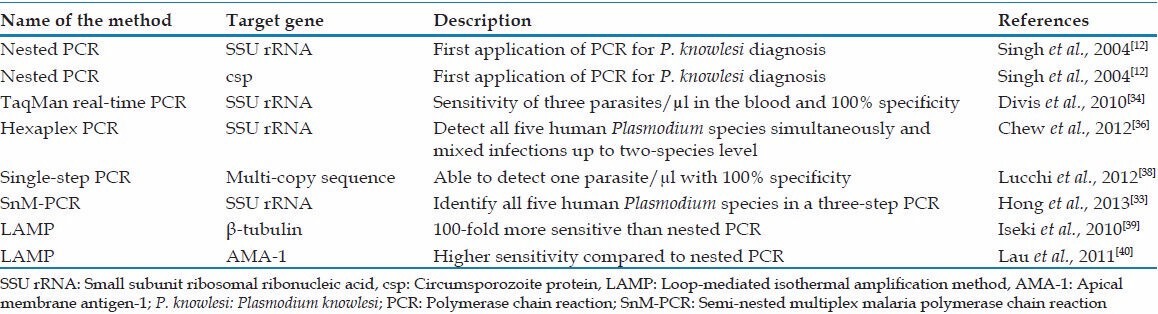
Two different loop-mediated isothermal amplification methods (LAMP) have been developed targeting species-specific β-tubulin gene[39] and apical membrane antigen-1 gene of P. knowlesi [Table 1].[40] It was shown that both LAMPs were specific and more sensitive than the nested-PCR targeting SSU rRNA.[39,40] The high sensitivity and specificity together with the high speed of LAMP and no need of sophisticated instruments like PCR makes LAMP a promising tool for diagnosing this human malaria parasite in the remote areas.
Beside PCR and LAMP, rapid diagnostic tests (RDTs) are very useful for epidemiological purposes in endemic areas where many people can be screened in a short period of time. However, RDTs are still not available to detect P. knowlesi. Commercial RDTs for malaria diagnosis use monoclonal antibodies that target one of three antigens namely histidine-rich protein 2 (HRP-2), Plasmodium lactate dehydrogenase (pLDH) and aldolase.[41] McCutchan et al. showed that P. knowlesi can react with the monoclonal antibodies targeting pLDH of P. falciparum and P. vivax possibly due to the highly similar amino acid sequences among these malaria parasites.[42] However, P. knowlesi did not react with HRP-2-based RDT, since HRP-2 is only expressed in P. falciparum.[43] Targeting other antigens which are abundant and well-conserved among human Plasmodium species may improve prompt detection of knowlesi malaria.[44]
APPLICATION OF MOLECULAR METHODS FOR EVALUATION OF DRUG RESISTANCE AND ITS SPREAD
After chloroquine resistance development in P. falciparum, several studies have been done to find the molecular background of the resistance with regard to gene mutation and copy number.[45,46,47] Recently, there have been some studies to evaluate antimalarial susceptibility of P. knowlesi.[48,49,50] Ex vivo studies on P. knowlesi isolates in Malaysian Borneo showed that these parasites are sensitive to artemisinins and chloroquine while they are less sensitive to mefloquine.[49] In another study, chloroquine resistance transporter and dihydrofolate reductase sequences of P. knowlesi clinical isolates in the Andaman and Nicobar Islands, India, were all found to be wild type[51] which these findings are consistent with Faith's findings.[49] Increased copy number of multidrug resistant gene 1 (mdr1) in P. falciparum is the most important determinant of resistance to mefloquine.[52] Although the copy number of P. knowlesi mdr1 was not reported in Faith's study,[49] less sensitivity of P. knowlesi to mefloquine may indicate an innate tolerance[49] or acquired tolerance under drug pressure since this parasite is not newly emerged[13] and has caused malaria in a great number of patients since 1996.[53]
Since P. knowlesi is a major cause of malaria in Malaysia[12] and large human population in Southeast Asia may be at risk of infection, it is worthy to continuously screen the drug susceptibility of this human malaria parasite especially in the mixed infections to better formulate an appropriate drug policy. The recent successful culture of P. knowlesi in human erythrocytes[54] pave the way for such drug screening and other in vitro studies on this human malaria parasite.
CONCLUSION
In the post-genomic era where the genome sequences of several Plasmodium species are available, molecular biological studies and genetic investigations should be combined effectively to epidemiological and clinical studies. Comparative genomics, molecular evolutionary analysis, population genetics and phylogenetic studies are some of the new research fields which were developed after the recent advances in Plasmodium genome. Human and mosquito vector genetics can influence the epidemiology of malaria in many potential aspects of ecological and evolutionary interaction.[55,56] There have been some studies to find the correlation between polymorphisms in specific genes and P. falciparum virulence.[57,58,59,60] In order to expand our knowledge on the pathogenesis of P. knowlesi which also can be fatal,[61] similar studies could be worthy.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, et al. Global malaria mortality between 1980 and 2010: A systematic analysis. Lancet. 2012;379:413–31. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 2.White NJ. Plasmodium knowlesi: The fifth human malaria parasite. Clin Infect Dis. 2008;46:172–3. doi: 10.1086/524889. [DOI] [PubMed] [Google Scholar]
- 3.Sabbatani S, Fiorino S, Manfredi R. The emerging of the fifth malaria parasite (Plasmodium knowlesi): A public health concern? Braz J Infect Dis. 2010;14:299–309. [PubMed] [Google Scholar]
- 4.Pain A, Böhme U, Berry AE, Mungall K, Finn RD, Jackson AP, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–6. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 7.Conway DJ. Molecular epidemiology of malaria. Clin Microbiol Rev. 2007;20:188–204. doi: 10.1128/CMR.00021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napier LE, Campbell HG. Observations on a Plasmodium infection which causes haemaglobinuria in certain species of monkey. Ind Med Gaz. 1932;67:246–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles R, Das Gupta BM. A study of monkey-malaria, and its experimental transmission to man. Ind Med Gaz. 1932;67:301–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Chin W, Contacos PG, Coatney GR, Kimball HR. A naturally acquited quotidian-type malaria in man transferable to monkeys. Science. 1965;149:865. doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- 11.Chin W, Contacos PG, Collins WE, Jeter MH, Alpert E. Experimental mosquito-transmission of Plasmodium knowlesi to man and monkey. Am J Trop Med Hyg. 1968;17:355–8. doi: 10.4269/ajtmh.1968.17.355. [DOI] [PubMed] [Google Scholar]
- 12.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 13.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–71. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. 2004;10:2211–3. doi: 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng OT, Ooi EE, Lee CC, Lee PJ, Ng LC, Pei SW, et al. Naturally acquired human Plasmodium knowlesi infection, Singapore. Emerg Infect Dis. 2008;14:814–6. doi: 10.3201/eid1405.070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luchavez J, Espino F, Curameng P, Espina R, Bell D, Chiodini P, et al. Human Infections with Plasmodium knowlesi, the Philippines. Emerg Infect Dis. 2008;14:811–3. doi: 10.3201/eid1405.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van den Eede P, Van HN, Van Overmeir C, Vythilingam I, Duc TN, Hung Le X, et al. Human Plasmodium knowlesi infections in young children in central Vietnam. Malar J. 2009;8:249. doi: 10.1186/1475-2875-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang N, Chang Q, Sun X, Lu H, Yin J, Zhang Z, et al. Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010;16:1476–8. doi: 10.3201/eid1609.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figtree M, Lee R, Bain L, Kennedy T, Mackertich S, Urban M, et al. Plasmodium knowlesi in human, Indonesian Borneo. Emerg Infect Dis. 2010;16:672–4. doi: 10.3201/eid1604.091624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khim N, Siv S, Kim S, Mueller T, Fleischmann E, Singh B, et al. Plasmodium knowlesi infection in humans, Cambodia, 2007-2010. Emerg Infect Dis. 2011;17:1900–2. doi: 10.3201/eid1710.110355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bronner U, Divis PC, Färnert A, Singh B. Swedish traveller with Plasmodium knowlesi malaria after visiting Malaysian Borneo. Malar J. 2009;8:15. doi: 10.1186/1475-2875-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Hellemond JJ, Rutten M, Koelewijn R, Zeeman AM, Verweij JJ, Wismans PJ, et al. Human Plasmodium knowlesi infection detected by rapid diagnostic tests for malaria. Emerg Infect Dis. 2009;15:1478–80. doi: 10.3201/eid1509.090358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ta TT, Salas A, Ali-Tammam M, Martínez Mdel C, Lanza M, Arroyo E, et al. First case of detection of Plasmodium knowlesi in Spain by real time PCR in a traveller from Southeast Asia. Malar J. 2010;9:219. doi: 10.1186/1475-2875-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link L, Bart A, Verhaar N, van Gool T, Pronk M, Scharnhorst V. Molecular detection of Plasmodium knowlesi in a Dutch traveler by real-time PCR. J Clin Microbiol. 2012;50:2523–4. doi: 10.1128/JCM.06859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehrhardt J, Trein A, Kremsner PG, Frank M. Plasmodium knowlesi and HIV co-infection in a German traveller to Thailand. Malar J. 2013;12:283. doi: 10.1186/1475-2875-12-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention (CDC) Simian malaria in a U.S. traveler – New York, 2008. MMWR Morb Mortal Wkly Rep. 2009;58:229–32. [PubMed] [Google Scholar]
- 27.Hoosen A, Shaw MT. Plasmodium knowlesi in a traveller returning to New Zealand. Travel Med Infect Dis. 2011;9:144–8. doi: 10.1016/j.tmaid.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Kuo MC, Chiang TY, Chan CW, Tsai WS, Ji DD. A case report of simian malaria, Plasmodium knowlesi, in a Taiwanese traveler from Palawan Island, the Philippines. Taiwan Epidemiol Bull. 2009;25:178–91. [Google Scholar]
- 29.Tanizaki R, Ujiie M, Kato Y, Iwagami M, Hashimoto A, Kutsuna S, et al. First case of Plasmodium knowlesi infection in a Japanese traveller returning from Malaysia. Malar J. 2013;12:128. doi: 10.1186/1475-2875-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallum MA, Peyton EL, Harrison BA, Wilkerson RC. Revision of the Leucosphyrus group of Anopheles (Cellia) (Diptera Culicidae) Rev Bras Entomol. 2005;49:1–152. [Google Scholar]
- 31.Lee KS, Cox-Singh J, Singh B. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 2009;8:73. doi: 10.1186/1475-2875-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imwong M, Tanomsing N, Pukrittayakamee S, Day NP, White NJ, Snounou G. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J Clin Microbiol. 2009;47:4173–5. doi: 10.1128/JCM.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Hong N, van den Eede P, Van Overmeir C, Vythilingham I, Rosanas-Urgell A, Vinh Thanh P, et al. A modified semi-nested multiplex malaria PCR (SnM-PCR) for the identification of the five human Plasmodium species occurring in Southeast Asia. Am J Trop Med Hyg. 2013;89:721–3. doi: 10.4269/ajtmh.13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Divis PC, Shokoples SE, Singh B, Yanow SK. A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malar J. 2010;9:344. doi: 10.1186/1475-2875-9-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calderaro A, Piccolo G, Gorrini C, Rossi S, Montecchini S, Dell'Anna ML, et al. Accurate identification of the six human Plasmodium spp. causing imported malaria, including Plasmodium ovale wallikeri and Plasmodium knowlesi. Malar J. 2013;12:321. doi: 10.1186/1475-2875-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chew CH, Lim YA, Lee PC, Mahmud R, Chua KH. Hexaplex PCR detection system for identification of five human Plasmodium species with an internal control. J Clin Microbiol. 2012;50:4012–9. doi: 10.1128/JCM.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goh XT, Lim YA, Vythilingam I, Chew CH, Lee PC, Ngui R, et al. Increased detection of Plasmodium knowlesi in Sandakan division, Sabah as revealed by PlasmoNex™. Malar J. 2013;12:264. doi: 10.1186/1475-2875-12-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lucchi NW, Poorak M, Oberstaller J, DeBarry J, Srinivasamoorthy G, Goldman I, et al. A new single-step PCR assay for the detection of the zoonotic malaria parasite Plasmodium knowlesi. PLoS One. 2012;7:e31848. doi: 10.1371/journal.pone.0031848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iseki H, Kawai S, Takahashi N, Hirai M, Tanabe K, Yokoyama N, et al. Evaluation of a loop-mediated isothermal amplification method as a tool for diagnosis of infection by the zoonotic simian malaria parasite Plasmodium knowlesi. J Clin Microbiol. 2010;48:2509–14. doi: 10.1128/JCM.00331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau YL, Fong MY, Mahmud R, Chang PY, Palaeya V, Cheong FW, et al. Specific, sensitive and rapid detection of human Plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J. 2011;10:197. doi: 10.1186/1475-2875-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.malERA Consultative Group on Diagnoses and Diagnostics. A research agenda for malaria eradication: Diagnoses and diagnostics. PLoS Med. 2011;8:e1000396. doi: 10.1371/journal.pmed.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCutchan TF, Piper RC, Makler MT. Use of malaria rapid diagnostic test to identify Plasmodium knowlesi infection. Emerg Infect Dis. 2008;14:1750–2. doi: 10.3201/eid1411.080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawai S, Hirai M, Haruki K, Tanabe K, Chigusa Y. Cross-reactivity in rapid diagnostic tests between human malaria and zoonotic simian malaria parasite Plasmodium knowlesi infections. Parasitol Int. 2009;58:300–2. doi: 10.1016/j.parint.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Hakimi H, Nguyen TT, Suganuma K, Masuda-Suganuma H, Angeles JM, Inoue N, et al. Development of Monoclonal Antibodies That Target 1-Cys Peroxiredoxin and Differentiate Plasmodium falciparum from P. vivax and P. knowlesi. Trop Med Health. 2013;41:55–9. doi: 10.2149/tmh.2012-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–6. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 46.Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–63. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 47.Hayton K, Su XZ. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr Drug Targets Infect Disord. 2004;4:1–10. doi: 10.2174/1568005043480925. [DOI] [PubMed] [Google Scholar]
- 48.Daneshvar C, Davis TM, Cox-Singh J, Rafa'ee MZ, Zakaria SK, Divis PC, et al. Clinical and parasitological response to oral chloroquine and primaquine in uncomplicated human Plasmodium knowlesi infections. Malar J. 2010;9:238. doi: 10.1186/1475-2875-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fatih FA, Staines HM, Siner A, Ahmed MA, Woon LC, Pasini EM, et al. Susceptibility of human Plasmodium knowlesi infections to anti-malarials. Malar J. 2013;12:425. doi: 10.1186/1475-2875-12-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barber BE, William T, Grigg MJ, Menon J, Auburn S, Marfurt J, et al. A prospective comparative study of knowlesi, falciparum, and vivax malaria in Sabah, Malaysia: High proportion with severe disease from Plasmodium knowlesi and Plasmodium vivax but no mortality with early referral and artesunate therapy. Clin Infect Dis. 2013;56:383–97. doi: 10.1093/cid/cis902. [DOI] [PubMed] [Google Scholar]
- 51.Tyagi RK, Das MK, Singh SS, Sharma YD. Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J Antimicrob Chemother. 2013;68:1081–8. doi: 10.1093/jac/dks508. [DOI] [PubMed] [Google Scholar]
- 52.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee KS, Cox-Singh J, Brooke G, Matusop A, Singh B. Plasmodium knowlesi from archival blood films: Further evidence that human infections are widely distributed and not newly emergent in Malaysian Borneo. Int J Parasitol. 2009;39:1125–8. doi: 10.1016/j.ijpara.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moon RW, Hall J, Rangkuti F, Ho YS, Almond N, Mitchell GH, et al. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci U S A. 2013;110:531–6. doi: 10.1073/pnas.1216457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–92. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niaré O, Markianos K, Volz J, Oduol F, Touré A, Bagayoko M, et al. Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science. 2002;298:213–6. doi: 10.1126/science.1073420. [DOI] [PubMed] [Google Scholar]
- 57.Ariey F, Hommel D, Le Scanf C, Duchemin JB, Peneau C, Hulin A, et al. Association of severe malaria with a specific Plasmodium falciparum genotype in French Guiana. J Infect Dis. 2001;184:237–41. doi: 10.1086/322012. [DOI] [PubMed] [Google Scholar]
- 58.Cramer JP, Mockenhaupt FP, Möhl I, Dittrich S, Dietz E, Otchwemah RN, et al. Allelic dimorphism of the erythrocyte binding antigen-175 (eba-175) gene of Plasmodium falciparum and severe malaria: Significant association of the C-segment with fatal outcome in Ghanaian children. Malar J. 2004;3:11. doi: 10.1186/1475-2875-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kun JF, Schmidt-Ott RJ, Lehman LG, Lell B, Luckner D, Greve B, et al. Merozoite surface antigen 1 and 2 genotypes and rosetting of Plasmodium falciparum in severe and mild malaria in Lambaréné, Gabon. Trans R Soc Trop Med Hyg. 1998;92:110–4. doi: 10.1016/s0035-9203(98)90979-8. [DOI] [PubMed] [Google Scholar]
- 60.Nielsen MA, Staalsoe T, Kurtzhals JA, Goka BQ, Dodoo D, Alifrangis M, et al. Plasmodium falciparum variant surface antigen expression varies between isolates causing severe and nonsevere malaria and is modified by acquired immunity. J Immunol. 2002;168:3444–50. doi: 10.4049/jimmunol.168.7.3444. [DOI] [PubMed] [Google Scholar]
- 61.Daneshvar C, Davis TM, Cox-Singh J, Rafa'ee MZ, Zakaria SK, Divis PC, et al. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis. 2009;49:852–60. doi: 10.1086/605439. [DOI] [PMC free article] [PubMed] [Google Scholar]


