Abstract
The IKKβ/NF-κB pathway is known to play an important role in inflammatory response and has also recently been implicated in the process of insulin resistance. We hypothesized that one or more variants in the IκBα gene (NFKBIA) or surrounding untranslated regions would be associated with insulin sensitivity (SI) in Hispanic-American families. We tested for association between 25 single-nucleotide polymorphisms (SNPs) in and near NFKBIA and SI in 981 individuals in 90 Hispanic-American families from the Insulin Resistance Atherosclerosis (IRAS) Family Study. SNP rs1951276 in the 3′ flanking region of NFKBIA was associated with SI in the San Antonio (SA) sample after adjusting for age, gender, and admixture (uncorrected P = 1.69 × 10−5; conservative Bonferroni correction P = 3.38 × 10−4). Subjects with at least one A allele for NFKBIA rs1951276 had ~29% lower SI compared to individuals homozygous for the G allele in the SA sample. Although not statistically significant, the effect was in the same direction in the San Luis Valley (SLV) sample alone (P = 0.348) and was significant in the combined SA and SLV samples (P = 5.37 × 10−4; presence of A allele associated with ~20% lower SI). In SA, when adjusted for subcutaneous adipose tissue area (SAT, cm2), the association was modestly attenuated (P = 1.25 × 10−3), but the association remained highly significant after adjustment for visceral adipose tissue area (VAT, cm2; P = 4.41 × 10−6). These results provide corroborating evidence that the NF-κB/IKKβ pathway may mediate obesity-induced insulin resistance in humans.
INTRODUCTION
Insulin resistance is a risk factor for type 2 diabetes (T2D) and cardiovascular disease. The prevalence of T2D has steadily risen in the United States and worldwide for the past several decades and is associated with significant morbidity and mortality. Increasing insulin resistance has been shown to strongly and consistently predict T2D (1–3) such that understanding the biological mechanisms of insulin resistance may provide insight to prevention strategies for T2D.
Several lines of evidence point to a relationship between inflammation and insulin resistance even if the nature of the relationship is unknown. Elevated serum levels of several proinflammatory cytokines have been shown to be associated with insulin resistance in epidemiologic studies (4,5). Obesity, a risk factor for insulin resistance, is also associated with chronic low-level inflammation (6). Furthermore, treatment of individuals with high doses of anti-inflammatory salicylates has long been known to decrease blood glucose levels in diabetic patients (7–11). However, until relatively recently, the mechanism of this action was poorly understood. Kopp and Ghosh (12) showed that aspirin and sodium salicylate inhibited the action of nuclear factor kappa B (NF-κB), a transcription factor which is upregulated by inflammatory stimuli and regulates the transcription of several inflammatory stimuli such as interleukin-6 and tumor necrosis factor-α. In addition, it has been shown that aspirin and sodium salicylates specifically inhibit the action of serine kinase IκB kinase-β (IKKβ), one of three kinases in the IKK complex that regulates NF-κB (13). As the NF-κB/IKKβ pathway is mediated by the inflammatory response, and because inhibition of the pathway improves blood glucose levels, Shoelson et al. (14) hypothesized that the NF-κB/IKKβ pathway might explain, in part, the relationship between insulin resistance and inflammation, particularly in the context of obesity.
Inhibitor of NF-κB alpha (IκBα) inhibits NF-κB by binding to one or more of its components, sequestering it in the cytoplasm until IκB is degraded after phosphorylation by IKKβ. Gao et al. (15) demonstrated that aspirin treatment prevents IκBα degradation in human muscle and a decrease in IκBα in human skeletal muscle is associated with fatty acid-induced insulin resistance (16). Despite the evidence that IκBα may be relevant to insulin resistance, no genetic studies have explored the potential for association between the IκBα gene (NFKBIA) and insulin resistance.
Given the importance of IκBα in regulating the NF-κB pathway, we investigated the relationship between several variants in NFKBIA and a measure of insulin sensitivity (SI) from a frequently sampled intravenous glucose tolerance test. We show that single-nucleotide polymorphism (SNP) rs1951276, located in the 3′ flanking region of NFKBIA, is associated with SI in a Hispanic-American sample. These results provide corroborating evidence that genetic variation the IKKβ/NF-κB pathway, and IκBα in particular, may mediate insulin resistance in humans.
METHODS AND PROCEDURES
Subject recruitment
Study design, recruitment, and phenotyping for the Insulin Resistance Atherosclerosis (IRAS) Family Study have been described in detail (17). Briefly, the IRAS Family Study is a multicenter study designed to investigate the genetic determinants of glucose homeostasis and adiposity. Probands who self-identified as Hispanic American or African American were first identified from the IRAS cohort study (18) and recruitment was supplemented with non-IRAS participants and their families. All families were recruited based on size and structure without regard to glucose tolerance or T2D status. Hispanic participants were recruited from two clinic sites, one in San Antonio (SA), Texas, and one in San Luis Valley (SLV), Colorado. There were a total of 1,268 Hispanic individuals from SA, Texas (60 families; 649 individuals), or the SLV, Colorado (30 families; 619 individuals) who had DNA available. For this study, a total of 1,147 individuals from 90 Hispanic families who had a frequently sampled intravenous glucose tolerance test were available from SA (586) or the SLV (561). All protocols were approved by the institutional review boards at each institution and all participants provided written informed consent prior to participation.
Clinical protocols
Clinic examination of participants included a frequently sampled intravenous glucose tolerance test using the reduced sampling protocol (19) as previously described (20). In addition to anthropometric measures of adiposity, abdominal fat area was measured using a computed tomography scan for abdominal adiposity measurements at the L3/L4 and L4/L5 vertebral regions for subcutaneous adipose tissue area (SAT) and visceral adipose tissue area (VAT) as previously described (17).
Genotyping
We initially genotyped 11 SNPs in and around NFKBIA (rs1951276, rs743228, rs7152826, rs3138056, rs3138055, rs696, rs8904, rs3138054, rs1967106, rs2233409, and rs3138053). SNPs were chosen based on linkage disequilibrium (LD) information from the HapMap Project (www.hapmap.org) and SeattleSNPs (http://pga.gs.washington.edu) resources and to capture regions implicated in association studies with other diseases. SNPs were selected from the CEU (population of northern and western European ancestry) population of HapMap and the European population of SeattleSNPs. To sample additional variability in the region of interest, additional SNPs were chosen for genotyping based upon LD structure in the YRI (Yoruba in Ibadan, Nigeria) samples from HapMap. At least one genotyped SNP had an r2 ≥ 0.65 with all known common (minor allele frequency >5%) variants in and near the gene (including 20 kilobases in the 3′ untranslated and flanking regions and 5 kilobases in the 5′ untranslated regions (UTR)) at the time of initial screening. Genotyping was performed on a MassARRAY system (Sequenom, San Diego, CA) as previously described (21). Based on 60 duplicate samples, the overall discrepancy rate was 0% and overall genotype success rate was >92%. Based on the results with the first 11 SNPs, we subsequently genotyped an additional 14 SNPs (rs8019505, rs12891443, rs3138045, rs4982270, rs2007960, rs17103274, rs8013309, rs1012919, rs17103282, rs8018407, rs8018193, rs2415290, rs8008601, rs762009, and rs4982271) so that at least one typed SNP had an r2 ≥ 0.90 with all known common variants in and near the gene (including 20 kilobases in the 5′ UTR).
Statistical analysis
The primary outcome for this study was SI, a measure of insulin sensitivity derived using minimal model (22) analysis of glucose and insulin data from the frequently sampled intravenous glucose tolerance (MINMOD Millennium, v5.18; MINMOD, Pasadena, CA).
Each SNP was checked for Mendelian inconsistencies using Pedcheck (23); inconsistencies that could not be resolved by inspection of genotype spectra were converted to missing. Genotype frequencies at each SNP were tested for consistency with Hardy–Weinberg Equilibrium proportions using the exact test based on the largest group of unrelated individuals using the PEDSTATS (24) package. Allele frequencies for each SNP were estimated using family data with the MENDEL program (v5.7) (25). LD measures (D′ and r2) were calculated in Haploview, v4.0 using the largest group of unrelated individuals (26).
Tests of association
We compared phenotypic means between clinic sites using a variance components model, which directly models the covariance among related individuals. Continuous outcomes SI, acute insulin response (AIR) and disposition index (DI) were natural log or square root transformed for analysis to best approximate the model assumptions (i.e., conditional multivariate normality and homogeneity of variance). Individuals with T2D were excluded (66 from SA and 41 from SLV). We tested for phenotypic differences between clinic sites using the variance components model. We tested for association between each SNP and SI using a likelihood ratio test under a variance components measured genotype approach as implemented in Simultaneous Oligogenic Linkage Analysis Routines (v.2.1.4) (27). For each clinic site separately, we tested for association using the likelihood ratio test under an additive model, unless the number of homozygous individuals for the minor allele was <10, where we tested under the dominant model (defined as presence of at least one copy of the minor allele). For SNPs that were highly associated with SI, we also tested for associations with other clinically relevant phenotypes such as AIR or DI.
For the first 11 SNPs we genotyped, each test was adjusted for (i) admixture (see below), age, and gender only, and (ii) admixture, age, gender, and BMI. Because SNPs rs696 and rs8904 were in nearly perfect LD (see Results), we tested for association with only rs696. Based on the results with the BMI adjustment, we conducted analyses with rs1951276, adjusting for VAT, SAT and VAT to SAT ratio (VSR). Finally, based on the results for the first 11 SNPs (rs1951276 in particular), for the next 14 SNPs that were subsequently genotyped, we adjusted for (i) admixture, age, and gender and (ii) admixture, age, gender, and VAT.
Admixture adjustment
The IRAS Family Study has genotyped 80 unlinked SNPs across the genome on a large fraction of IRAS Family Study participants. These markers were chosen as ancestry informative markers (K. Taylor, personal communication) for the purposes of admixture adjustment. Based on a principal components analysis of these 80 markers, the second principal component explained a significant proportion of the genetic variance in the Hispanic sample and represents the proportion of European admixture for each individual. We adjusted for admixture by including this principal component as a covariate in our Simultaneous Oligogenic Linkage Analysis Routines models.
RESULTS
We report results from 499 individuals in the SA sample and 482 individuals in the SLV sample with complete frequently sampled intravenous glucose tolerance tests who were successfully genotyped on at least one NFKBIA SNP and the 80 ancestry informative markers. The number of individuals per family ranged from 3 to 48 with a mean of 21. Descriptive characteristics for each clinic sample are shown in Table 1. Individuals from the SLV sample tended to have lower BMI (P = 5.48 × 10−5) and better glucose homeostasis profiles compared to individuals from the SA sample (higher SI (P = 0.0040) and DI (P = 9.79 × 10−7); lower AIR (P = 0.0357) and glucose effectiveness (P = 5.19 × 10−15)). The second principal component explained 5.1% of the genetic variation in the combined Hispanic samples and differed markedly between the two clinic sites (P = 1.03 × 10−13).
Table 1.
Study characteristics by site
| San Antonio (n = 499) | San Luis Valley (n = 482) | P value | |
|---|---|---|---|
| Male/female | 195/304 | 208/274 | 0.2169 |
| Age (years) | 40.33 (14.13) | 41.47 (13.28) | 0.3146 |
| Insulin sensitivity (×10−3 min−1 pmol/l) | 3.25 (3.05) | 3.92 (3.18) | 0.0041 |
| BMI (kg/m2) | 29.61 (6.17) | 27.08 (5.23) | 5.48 × 10−5 |
| Visceral adipose area (VAT) (cm2) | 112.45 (59.09) | 102.65 (54.38) | 0.0040 |
| Subcutaneous adipose area (SAT) (cm2) | 346.13 (158.04) | 316.53 (145.12) | 0.0629 |
| Acute insulin response (AIR) pmol/l × 10 min | 722.57 (651.39) | 812.71 (667.38) | 0.0357 |
| Glucose effectiveness (SG) min−1 | 0.018 (0.008) | 0.023 (0.009) | 5.19 × 10−15 |
| Disposition index (SI × AIR) | 1,770.66 (1,552.64) | 2,639.74 (2,408.21) | 9.79 × 10−7 |
| Principal componenta | 0.59 (0.15) | 0.50 (0.09) | 1.03 × 10−13 |
Values are mean (s.d.), except male/female.
Principal component score based on 80 Hispanic ancestry informative genetic markers.
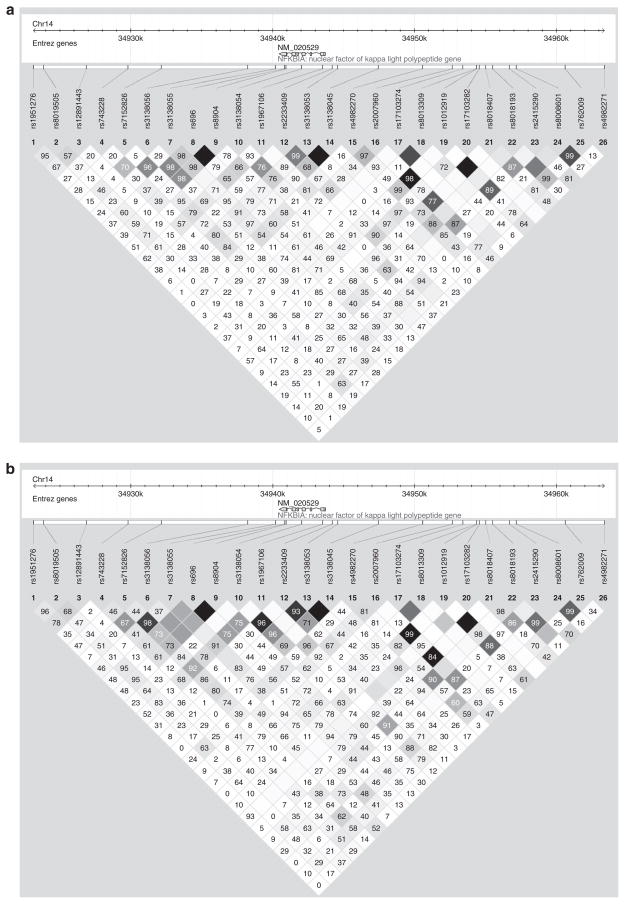
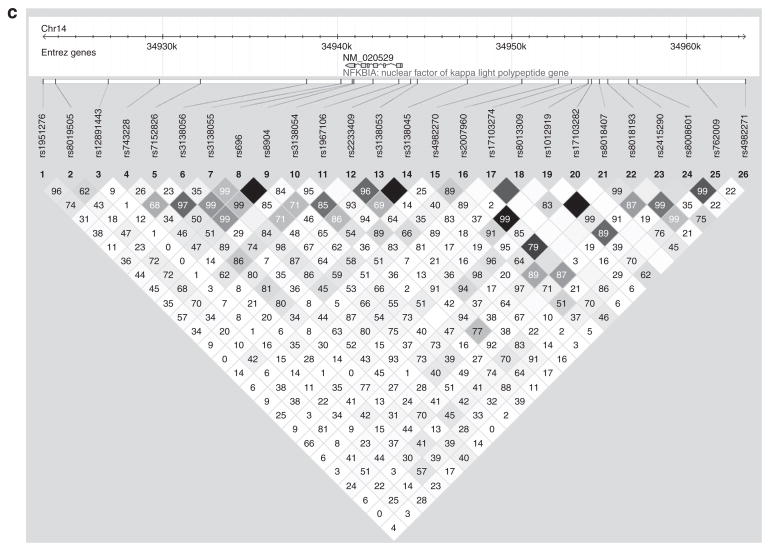
NFKBIA is located on chromosome 14, is only 3.3 kilobases long, and has six exons. Characteristics of all 25 genotyped SNPs are shown in Supplementary Table S1 online. Minor allele frequencies are similar between the two samples for many of the SNPs, but show marked differences for SNPs rs8019505, rs7152826, rs3138055, rs696, rs8904, and rs3138053. In addition, for SNPs rs3138056 and rs4982270, the clinic sites have opposite minor alleles. Figure 1 shows the pair-wise LD for all 25 SNPs in and near NFKBIA for each sample. LD is displayed as pair-wise r2 values, where white indicates r2 = 0, varying shades of gray indicate 0 < r2 < 1, and black indicates r2 = 1. Numeric values are pair-wise D′ values × 100. Patterns of LD were similar in both samples. As SNPs rs696 and rs8904 were in near-perfect LD (r2 = 0.96 in SA, r2 = 1.0 in SLV), we tested for association with rs696 only.
Figure 1.
Pair-wise LD for all 25 SNPs in and around the NFKBIA gene in (a) San Antonio, (b) San Luis Valley, and (c) the Combined Hispanic sample. LD is displayed as pair-wise r2 values, where white indicates r2 = 0, varying shades of gray indicate 0 < r2 < 1, and black indicates that r2 = 1. Numeric values are pair-wise D′ values × 100.
Results of the tests for association with each of the SNPs and SI in the SA sample, adjusted for admixture, age and gender, are shown in Table 2. Among the first 11 SNPs typed, rs1951276 was highly associated with SI (P = 3.52 × 10−5). In addition, rs7152826 was nominally associated with SI (P = 0.024). The results were largely similar after adjustment for BMI (Supplementary Table S2 online); the association between rs1951276 and SI remained significant (P = 2.32 × 10−3). Even with a very conservative Bonferroni adjustment for the 20 tests performed (10 SNPs, two covariate adjustments), the association between rs1951276 and SI remains significant (P = 7.04 × 10−4) with admixture, age, and gender as covariates.
Table 2.
Association of NFKBIA SNPs with SI in San Antonio sample
| SNP | MAF | Genotypic mean (s.d.) n
|
P valuea | ||
|---|---|---|---|---|---|
| 11 | 12 | 22 | |||
| rs1951276b | 0.224 | 2.80 (1.85) n = 279 | 2.03 (1.82) n = 166 | 1.90 (1.85) n =15 | 3.52 × 10−5 |
| rs8019505 | 0.364 | 2.72 (1.73) n = 207 | 2.57 (1.86) n =214 | 2.27 (1.86) n = 60 | 0.127 |
| rs12891443 | 0.455 | 2.37 (1.88) n = 142 | 2.56 (1.89) n = 234 | 2.35 (1.68) n = 106 | 0.519 |
| rs743228b | 0.410 | 2.54 (1.86) n =176 | 2.57 (1.91) n = 217 | 2.27 (1.72) n = 91 | 0.861 |
| rs7152826b | 0.468 | 2.41 (1.87) n = 125 | 2.37 (1.88) n = 233 | 2.83 (1.77) n = 131 | 0.024 |
| rs3138056b | 0.490 | 2.51 (1.78) n = 129 | 2.65 (1.88) n = 242 | 2.13 (1.83) n = 109 | 0.150 |
| rs3138055b | 0.372 | 2.63 (1.84) n = 203 | 2.34 (1.88) n = 221 | 2.39 (1.83) n = 46 | 0.055 |
| rs696b,c | 0.314 | 2.46 (1.79) n = 193 | 2.51 (1.87) n = 226 | 2.29 (1.95) n = 64 | 0.438 |
| rs3138054b | 0.107 | 2.44 (1.88) n = 387 | 2.58 (1.72) n = 89 | 2.82 (1.96) n = 6 | 0.804d |
| rs1957106b | 0.202 | 2.43 (1.83) n = 295 | 2.52 (1.85) n = 151 | 2.41 (2.03) n = 34 | 0.176 |
| rs2233409b | 0.130 | 2.49 (1.88) n = 359 | 2.30 (1.76) n = 118 | 4.11 (1.28) n = 3 | 0.745d |
| rs3138053b | 0.219 | 2.45 (1.92) n = 293 | 2.57 (1.80) n = 178 | 2.46 (1.55) n = 17 | 0.731 |
| rs3138045 | 0.185 | 2.44 (1.89) n = 322 | 2.55 (1.79) n = 153 | 2.27 (1.56) n = 13 | 0.923 |
| rs4982270 | 0.476 | 2.15 (1.90) n = 152 | 2.44 (1.78) n = 232 | 2.70 (1.91) n = 96 | 0.043 |
| rs2007960 | 0.252 | 2.09 (1.78) n = 292 | 2.23 (1.77) n = 164 | 2.66 (1.88) n = 29 | 0.021 |
| rs17103274 | 0.081 | 2.45 (1.85) n = 414 | 2.77 (1.88) n = 77 | 1.37 (2.37) n = 3 | 0.879d |
| rs8013309 | 0.119 | 2.46 (1.84) n = 372 | 2.64 (1.92) n = 110 | 1.96 (1.79) n = 9 | 0.662d |
| rs1012919 | 0.202 | 2.59 (1.87) n = 328 | 2.25 (1.83) n = 134 | 2.30 (1.78) n = 20 | 0.083 |
| rs17103282 | 0.033 | 2.54 (1.86) n = 450 | 2.01 (1.85) n = 36 | e | 0.543d |
| rs8018407 | 0.272 | 2.54 (1.87) n = 289 | 2.42 (1.81) n =165 | 2.04 (1.79) n = 27 | 0.214 |
| rs8018193 | 0.308 | 2.31 (1.84) n = 210 | 2.58 (1.8) n = 214 | 2.86 (2.05) n = 60 | 0.022 |
| rs2415290 | 0.161 | 2.21 (1.70) n = 364 | 2.13 (1.84) n = 110 | 2.58 (1.85) n = 9 | 0.024d |
| rs8008601 | 0.483 | 2.38 (1.90) n = 113 | 2.48 (1.82) n = 259 | 2.50 (1.85) n = 109 | 0.205 |
| rs762009 | 0.392 | 2.43 (1.80) n = 167 | 2.52 (1.88) n = 254 | 2.31 (1.86) n = 59 | 0.241 |
| rs4982271 | 0.391 | 2.61 (1.95) n = 214 | 2.37 (1.78) n = 208 | 2.51 (1.80) n = 70 | 0.265 |
All P values are adjusted for admixture, age, and gender. Mean values shown are the unadjusted genotypic means (s.d.) for SI. n is the number of individuals with that genotype. Nominally significant P values are in boldface.
P values are for the additive model unless indicated.
First 11 SNPs typed (r2 ≥ 0.65 with untyped SNPs). 14 SNPs with no superscript were typed after results from first 11 obtained (r2 ≥ 0.90 with untyped SNPs).
SNPs rs696 and rs8904 were in near-perfect LD (San Antonio: r2 = 0.963; San Luis Valley: r2= 1.00). Only SNP rs696 was tested for association with SI.
P values are for the dominant model because there were <10 minor allele homozygotes.
There were no minor allele homozygotes for this SNP.
Results of the tests for association with each of the SNPs and SI in the SLV sample are shown in Table 3. In contrast to the SA sample results, when adjusting for admixture, age, and gender, or admixture, age, gender, and BMI, none of the SNPs in the SLV sample were significantly associated with SI. Complete results for the adjustment of age, gender, and BMI in the SLV sample are in Supplementary Table S2 online.
Table 3.
Association of NFKBIA SNPs with SI in San Luis Valley sample
| SNP | MAF | Genotypic mean (s.d.) N
|
P valuea | ||
|---|---|---|---|---|---|
| 11 | 12 | 22 | |||
| rs1951276b | 0.277 | 3.17 (1.89) n = 258 | 2.86 (1.86) n =167 | 3.01 (2.00) n = 37 | 0.521 |
| rs8019505 | 0.313 | 3.05 (1.91) n = 239 | 3.06 (1.86) n = 187 | 3.13 (1.95) n = 44 | 0.803 |
| rs12891443 | 0.468 | 2.95 (1.88) n = 132 | 3.04 (1.86) n = 247 | 3.12 (2.00) n = 93 | 0.486 |
| rs743228b | 0.442 | 3.05 (1.98) n = 134 | 2.95 (1.89) n = 244 | 3.30 (1.79) n = 92 | 0.302 |
| rs7152826b | 0.386 | 3.15 (1.84) n = 177 | 2.91 (1.94) n = 254 | 3.44 (1.87) n = 48 | 0.215 |
| rs3138056b | 0.473 | 2.88 (1.99) n = 135 | 3.15 (1.87) n = 235 | 3.03 (1.75) n = 99 | 0.131 |
| rs3138055b | 0.480 | 3.05 (1.85) n = 122 | 2.81 (1.92) n = 256 | 3.56 (1.82) n = 91 | 0.889 |
| rs696b,c | 0.272 | 3.05 (1.89) n = 251 | 3.07 (1.89) n = 190 | 2.79 (1.85) n = 29 | 0.431 |
| rs3138054b | 0.107 | 3.07 (1.90) n = 398 | 2.90 (1.84) n = 72 | 3.75 (1.21) n = 1 | 0.951d |
| rs1957106b | 0.201 | 3.06 (1.87) n = 282 | 3.07 (1.89) n = 165 | 2.46 (2.08) n = 20 | 0.755 |
| rs2233409b | 0.135 | 3.17 (1.88) n = 368 | 2.57 (1.91) n = 99 | 3.12 (1.29) n = 3 | 0.117d |
| rs3138053b | 0.182 | 3.11 (1.86) n = 337 | 3.02 (2.00) n = 125 | 3.18 (1.54) n = 9 | 0.992d |
| rs3138045 | 0.146 | 3.06 (1.86) n = 359 | 2.99 (2.04) n = 106 | 3.18 (1.54) n = 9 | 0.663d |
| rs4982270 | 0.468 | 2.91 (1.82) n = 128 | 3.27 (1.92) n = 237 | 2.72 (1.92) n = 104 | 0.785 |
| rs2007960 | 0.257 | 2.58 (1.93) n = 274 | 3.17 (1.81) n = 172 | 3.03 (1.94) n = 31 | 0.857 |
| rs17103274 | 0.053 | 3.10 (1.89) n = 432 | 2.71 (1.79) n = 47 | e | 0.974d |
| rs8013309 | 0.088 | 3.13 (1.89) n = 394 | 2.70 (1.88) n = 83 | e | 0.559d |
| rs1012919 | 0.230 | 3.01 (1.96) n = 290 | 3.22 (1.77) n = 158 | 3.08 (1.94) n = 19 | 0.234 |
| rs17103282 | 0.026 | 3.10 (1.89) n = 443 | 2.40 (1.86) n = 26 | e | 0.304d |
| rs8018407 | 0.276 | 3.03 (1.96) n = 270 | 3.21 (1.77) n = 169 | 2.72 (1.85) n = 31 | 0.426 |
| rs8018193 | 0.342 | 3.00 (1.90) n = 208 | 3.22 (1.87) n = 208 | 2.70 (1.94) n = 62 | 0.918 |
| rs2415290 | 0.179 | 3.22 (2.03) n = 332 | 3.22 (1.78) n = 135 | 2.98 (1.93) n = 10 | 0.443 |
| rs8008601 | 0.491 | 3.36 (1.90) n = 125 | 2.98 (1.89) n = 237 | 2.81 (1.89) n = 110 | 0.596 |
| rs762009 | 0.395 | 3.17 (1.96) n = 176 | 3.06 (1.91) n = 225 | 2.99 (1.86) n = 68 | 0.727 |
| rs4982271 | 0.302 | 2.87 (1.94) n = 222 | 3.34 (1.83) n = 198 | 2.83 (1.88) n = 53 | 0.455 |
All P values are adjusted for admixture, age, and gender. Mean values shown are the unadjusted genotypic means (s.d.) for SI. n is the number of individuals with that genotype. Nominally significant P values are in boldface.
P values are for the additive model unless indicated.
First 11 SNPs typed (r2 ≥ 0.65 with untyped SNPs). 14 SNPs with no superscript were typed after the results from first 11 obtained (r2 ≥ 0.90 with untyped SNPs).
SNPs rs696 and rs8904 were in near-perfect LD (San Antonio: r2 = 0.963; San Luis Valley: r2 = 1.00). Only SNP rs696 was tested for association with SI.
P values are for the dominant model because there were <10 minor allele homozygotes.
There were no minor allele homozygotes for this SNP.
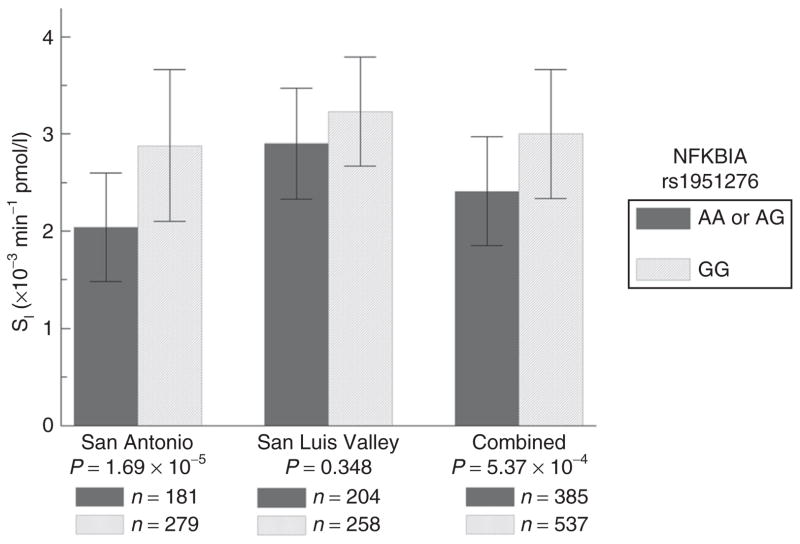
As the test for association between rs1951276 was significant in SA, we tested for association under dominant and recessive models. Under a dominant model, the evidence for association was slightly increased (P = 1.69 × 10−5), and inspection of the genotypic means indicated a dominant pattern. Although still not significant in the SLV sample under a dominant model (P = 0.348), the genotypic means at rs1951276 show the same pattern, and the test for association in the combined sample was significant (P = 5.37 × 10−4, adjusted for admixture, age, gender, and site). This relationship was slightly attenuated after adjustment for BMI (P = 2.42 × 10−3).
Because the association between rs1951276 was attenuated after adjustment for BMI in SA and the combined sample, we examined whether more specific measures of adiposity resulted in a similar reduction in the evidence for association with SI. In three different models, we adjusted for admixture, age, gender and either VAT, SAT, or VSR (Table 4). Similar to adjustment for BMI, adjustment for SAT in SA attenuated the evidence for association (SAT P = 1.25 × 10−3). However, adjustment for VAT and VSR resulted in slightly increased evidence (VAT P= 4.41 × 10−6; VSR = 9.10 × 10−6).
Table 4.
P Value for test of association between rs1951276 and SI under dominant model
| Adiposity measure | Clinic
|
||
|---|---|---|---|
| SA | SLV | Combineda | |
| None | 1.69 × 10−5 | 0.348 | 5.37 × 10−4 |
| BMI | 9.82 × 10−4 | 0.202 | 2.42 × 10−3 |
| VAT | 4.41 × 10−6 | 0.716 | 1.17 × 10−3 |
| SAT | 1.25 × 10−3 | 0.268 | 5.21 × 10−3 |
| VSR | 9.10 × 10−6 | 0.279 | 2.58 × 10−4 |
P values are adjusted for admixture, age, and gender only, or one of four measures of adiposity.
SA, San Antonio; SAT, subcutaneous adipose tissue; SLV, San Luis Valley; VAT, visceral adipose tissue; VSR, VAT to SAT ratio.
Also adjusted for site.
Figure 2 shows the association between rs1951276 and SI in the SA, SLV and combined samples. For SNP rs1951276 in the SA sample, presence of at least one copy of the minor allele A was associated with a ~29% reduction in mean SI. In the SLV sample, presence of at least one copy of the minor allele was associated with a ~10% reduction (not significant) in mean SI. Presence of at least one copy of the A allele was associated with ~20% reduction in mean SI in the combined sample. The percent increase was similar when adjusting for admixture, age, and gender only, or admixture, age, gender, and any of the adiposity covariates.
Figure 2.
Association with SI and NFKBIA rs1951276 in the IRAS Family Study. Values shown are the genotype specific means for NFKBIA rs1951276, adjusted for admixture, age, and gender, where rs1951276 is assumed to follow a dominant model.
We genotyped additional SNPs throughout the NFKBIA region so that each SNP with minor allele frequency >0.05 in the HapMap had an r2 of at least 0.90 with at least one genotyped SNP. As the most significant results for rs1951276 were obtained with either no adiposity adjustment or with adjustment for VAT, we tested for association with each of the new SNPs and either admixture, age and gender only or admixture, age, gender, and VAT. In the SA sample, after adjustment for admixture, age and gender, SNP rs4982270 (P = 0.043), rs2007960 (P = 0.021), rs8018193 (P = 0.022), and rs2415290 (dominant P = 0.024) were all nominally associated with SI (Table 2). After further adjustment for VAT, rs4982270 (P = 2.87 × 10−3), rs2007960 (P = 5.30 × 10−3), rs1012919 (P = 0.049), rs8018193 (P = 0.012), and rs2415290 (dominant P = 0.042) were all nominally associated with SI (Supplementary Table S3 online). None of the SNPs were associated with SI in the SLV sample.
Based on the strong association of rs1951276 with SI, we also tested whether this SNP was associated with other phenotypes under a dominant model, adjusting for admixture, age, and gender. In the SA sample, rs1951276 was strongly associated with BMI (P = 6.47 × 10−3), fasting insulin (P = 1.09 × 10−3), and 180 min insulin (5.37 × 10−5) and nominally associated with AIR (P = 0.048), DI (P = 0.030), and SAT (P = 0.018). Rs1951276 was not associated with fasting glucose, 180 min glucose, glucose effectiveness, VAT, or VSR in the SA sample. None of the phenotypes were associated with rs1951276 in the SLV sample. In the combined sample, additionally adjusted for clinic site, rs1951276 was associated with fasting insulin (P = 4.78 × 10−3), 180 min insulin (P = 3.46 × 10−3), and DI (P = 7.84 × 10−3).
DISCUSSION
We found a strong association between rs1951276 in the 3′ flanking of NFKBIA and SI in a sample of Hispanic-American families from SA, Texas. Although not significant in another sample of Hispanic-American families from the SLV, Colorado, the genotypic means showed the same pattern as those in the SA sample, and the association was significant in the combined samples. Having at least one copy of the A allele at rs1951276 was associated with ~29% lower SI in the SA sample alone, and 20% lower SI in the combined SA and SLV samples.
There are several possible explanations for the differential evidence for association in the two samples even though the pattern of genotypic means is very similar between the two groups. First, principle components analysis of a limited number of ancestry informative markers indicates a significant difference in the level of European admixture between the two samples, which is consistent with other studies of admixture in similar populations. Another admixture analysis in the SLV suggests a large proportion of “Spanish” (~60%) and less Native American (~30%) admixture (28). In contrast, much higher estimates of Native American admixture for Southwestern Hispanics have been reported (29). The results of the association tests were largely unchanged after adjustment for admixture, however. Second, the SLV is a high-altitude (~7,500 feet above sea level) rural setting whereas SA is an urban setting. Third, the two samples also differ in their adiposity and glucose homeostasis profiles. Individuals from the SLV sample tend to be leaner, (lower BMI, SAT, and VAT), and have better glucose homeostasis profiles (higher SI, AIR, DI) than those in the SA sample (see Table 1). Fourth, even though the SA and SLV samples have approximately the same number of individuals, the SLV sample has half the number of families as the SA sample. Thus, the much larger families in the SLV sample result in a lower effective sample size than the SA effective sample size. Finally, we cannot rule out the possibility of a false-positive result in the SA sample.
Although the NF-κB/IKKβ pathway appears to be particularly relevant for obesity-related insulin resistance (30), it is not known whether different adipose tissue depots exhibit different characteristics related to NF-κB/IKKβ activity. In our study, the evidence for association between rs1951276 and SI in the SA sample differed depending on which measure of adiposity we used as an adjustment. Specifically, when adjusted for BMI or SAT area, the association between rs1951276 was attenuated. However, the evidence for association was essentially unchanged after adjustment for VAT area or the ratio of visceral to SAT area, suggesting that the role of NFKBIA may differ depending on adipose tissue depot. Unfortunately, sample sizes were not sufficient to allow meaningful interpretation of analyses stratified by median BMI, SAT, or VAT that might more clearly address this question.
Rs1951276 is in the 3′ flanking region of NFKBIA, the gene for IκBα, and the potential functional role for this SNP or a functional variant in LD with rs1951276 is unknown. There are no known variants within 20 kilobases in either direction of rs1951276 with an r2 ≥ 0.31. It is possible that an unknown functional 3′ UTR or flanking variant affects post-translational modification of IκBα mRNA, affecting its quality or quantity (31). A reduction of IκBα is associated with fatty acid-induced insulin resistance (16), and there are at least two mechanisms by which altered or reduced IκBα might result in insulin resistance. As described above, inhibition of IKKβ has been shown to reduce SI. As IKKβ phosphorylates IκBα (32) and may similarly phosphorylate insulin receptor substrate-1/2 (33,34), IκBα may regulate IKKβ by acting as a competing substrate. Alternatively, a reduction in IκBα could result in increased translocation of NF-κB into the nucleus, which would increase transcription of several inflammatory cytokines associated with insulin resistance, such as tumor necrosis factor-α.
No other studies have examined the role of polymorphisms in NFKBIA and SI. However, polymorphisms in the 3′ UTR and promoter regions have been associated with Crohn’s disease (35), multiple sclerosis (36), and severe carotid artery disease (37), which like insulin resistance, have been associated with chronic inflammation (38,39). Although we saw nominal association between SI and some of the SNPs identified in these other studies, none of the signals were as strong as with rs1951276. None of these other studies typed rs1951276, so we do not know whether similar association would have been seen with this particular SNP. Because the LD in this region is fairly weak, future studies will further examine the region using sequencing to identify potential functional polymorphisms.
In conclusion, we have identified a variant in the 3′ intravenous region of NFKBIA that is associated with a 20–29% reduction in SI as measured by a frequently sampled glucose tolerance test in Hispanic Americans. These results corroborate other evidence that the NF-κB/IKKβ pathway may mediate obesity-induced insulin resistance in humans.
Supplementary Material
Acknowledgments
T.E.F. was supported by an American Diabetes Association Junior Faculty Award. The IRAS Family Study (IRASFS) is supported by NIH Grants HL-60944-02, HL-61210-02, HL-61019-02, HL-60894, and HL-60931-02. We thank the families who volunteered to participate in the IRASFS.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
DISCLOSURE
The authors declared no conflict of interest.
References
- 1.Lillioja S, Mott DM, Spraul M, et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med. 1993;329:1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 2.Haffner SM, Miettinen H, Gaskill SP, Stern MP. Decreased insulin secretion and increased insulin resistance are independently related to the 7-year risk of NIDDM in Mexican-Americans. Diabetes. 1995;44:1386–1391. doi: 10.2337/diab.44.12.1386. [DOI] [PubMed] [Google Scholar]
- 3.Weyer C, Bogardus C, Pratley RE. Metabolic characteristics of individuals with impaired fasting glucose and/or impaired glucose tolerance. Diabetes. 1999;48:2197–2203. doi: 10.2337/diabetes.48.11.2197. [DOI] [PubMed] [Google Scholar]
- 4.Yudkin JS, Yajnik CS, Mohamed-Ali V, Bulmer K. High levels of circulating proinflammatory cytokines and leptin in urban, but not rural, Indians. A potential explanation for increased risk of diabetes and coronary heart disease. Diabetes Care. 1999;22:363–364. doi: 10.2337/diacare.22.2.363. [DOI] [PubMed] [Google Scholar]
- 5.Festa A, D’Agostino R, Howard G, et al. Inflammation and microalbuminuria in nondiabetic and type 2 diabetic subjects: The Insulin Resistance Atherosclerosis Study. Kidney Int. 2000;58:1703–1710. doi: 10.1046/j.1523-1755.2000.00331.x. [DOI] [PubMed] [Google Scholar]
- 6.Hotamisligil GS. Inflammation, TNF-alpha, and insulin resistance. In: LeRoith DTS, Olefsky JM, editors. Diabetes Mellitus: A Fundamental and Clinical Text. 3. Lippincott, Williams, and Wilkins; New York: 2003. pp. 953–962. [Google Scholar]
- 7.Ebstein W. Zur Therapie des Diabetes Mellitus, Insbesondere Uber Die Anwendeng der Salicylauren Natron Bei Demselben. Berlin Lin Wochnschrift. 1876;13:337–340. [Google Scholar]
- 8.Williamson RT. On the Treatment of Glyosuria and Diabetes Mellitus with Sodium Salicylate. BMJ. 1901;1:760–762. doi: 10.1136/bmj.1.2100.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid J, MacDougall AI, Andrews MM. Aspirin and diabetes mellitus. Br Med J. 1957;2:1071–1074. doi: 10.1136/bmj.2.5053.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilgore S. The Influence of Salicylate on Hyperglycemia. Diabetes. 1960;9:392–393. [Google Scholar]
- 11.Baron SH. Salicylates as hypoglycemic agents. Diabetes Care. 1982;5:64–71. doi: 10.2337/diacare.5.1.64. [DOI] [PubMed] [Google Scholar]
- 12.Kopp E, Ghosh S. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- 13.Yin MJ, Yamamoto Y, Gaynor RB. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature. 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- 14.Shoelson SE, Lee J, Yuan M. Inflammation and the IKK beta/I kappa B/ NF-kappa B axis in obesity- and diet-induced insulin resistance. Int J Obes Relat Metab Disord. 2003;27(Suppl 3):S49–S52. doi: 10.1038/sj.ijo.0802501. [DOI] [PubMed] [Google Scholar]
- 15.Gao Z, Zuberi A, Quon MJ, Dong Z, Ye J. Aspirin inhibits serine phosphorylation of insulin receptor substrate 1 in tumor necrosis factor-treated cells through targeting multiple serine kinases. J Biol Chem. 2003;278:24944–24950. doi: 10.1074/jbc.M300423200. [DOI] [PubMed] [Google Scholar]
- 16.Itani SI, Ruderman NB, Schmieder F, Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkappaB-alpha. Diabetes. 2002;51:2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- 17.Henkin L, Bergman RN, Bowden DW, et al. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13:211–217. doi: 10.1016/s1047-2797(02)00412-x. [DOI] [PubMed] [Google Scholar]
- 18.Wagenknecht LE, Mayer EJ, Rewers M, et al. The insulin resistance atherosclerosis study (IRAS) objectives, design, and recruitment results. Ann Epidemiol. 1995;5:464–472. doi: 10.1016/1047-2797(95)00062-3. [DOI] [PubMed] [Google Scholar]
- 19.Steil GM, Volund A, Kahn SE, Bergman RN. Reduced sample number for calculation of insulin sensitivity and glucose effectiveness from the minimal model. Suitability for use in population studies. Diabetes. 1993;42:250–256. doi: 10.2337/diab.42.2.250. [DOI] [PubMed] [Google Scholar]
- 20.Rich SS, Bowden DW, Haffner SM, et al. Insulin Resistance Atherosclerosis Study. Family Study. Identification of quantitative trait loci for glucose homeostasis: the Insulin Resistance Atherosclerosis Study (IRAS) Family Study. Diabetes. 2004;53:1866–1875. doi: 10.2337/diabetes.53.7.1866. [DOI] [PubMed] [Google Scholar]
- 21.Buetow KH, Edmonson M, MacDonald R, et al. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci USA. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergman RN, Finegood DT, Ader M. Assessment of insulin sensitivity in vivo. Endocr Rev. 1985;6:45–86. doi: 10.1210/edrv-6-1-45. [DOI] [PubMed] [Google Scholar]
- 23.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 25.Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonilla C, Parra EJ, Pfaff CL, et al. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet. 2004;68:139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 29.Allard MW, Polanskey D, Wilson MR, Monson KL, Budowle B. Evaluation of variation in control region sequences for Hispanic individuals in the SWGDAM mtDNA data set. J Forensic Sci. 2006;51:566–573. doi: 10.1111/j.1556-4029.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 30.Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132:2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 31.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 32.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 33.Kim JK, Kim YJ, Fillmore JJ, et al. Prevention of fat-induced insulin resistance by salicylate. J Clin Invest. 2001;108:437–446. doi: 10.1172/JCI11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, Hwang D, Bataille F, et al. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J Biol Chem. 2002;277:48115–48121. doi: 10.1074/jbc.M209459200. [DOI] [PubMed] [Google Scholar]
- 35.Klein W, Tromm A, Folwaczny C, et al. A polymorphism of the NFKBIA gene is associated with Crohn’s disease patients lacking a predisposing allele of the CARD15 gene. Int J Colorectal Dis. 2004;19:153–156. doi: 10.1007/s00384-003-0531-y. [DOI] [PubMed] [Google Scholar]
- 36.Miterski B, Böhringer S, Klein W, et al. Inhibitors in the NFkappaB cascade comprise prime candidate genes predisposing to multiple sclerosis, especially in selected combinations. Genes Immun. 2002;3:211–219. doi: 10.1038/sj.gene.6363846. [DOI] [PubMed] [Google Scholar]
- 37.Carlson CS, Heagerty PJ, Nord AS, et al. TagSNP evaluation for the association of 42 inflammation loci and vascular disease: evidence of IL6, FGB, ALOX5, NFKBIA, and IL4R loci effects. Hum Genet. 2007;121:65–75. doi: 10.1007/s00439-006-0289-8. [DOI] [PubMed] [Google Scholar]
- 38.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 39.Clarkson AN, Rahman R, Appleton I. Inflammation and autoimmunity as a central theme in neurodegenerative disorders: fact or fiction? Curr Opin Investig Drugs. 2004;5:706–713. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.