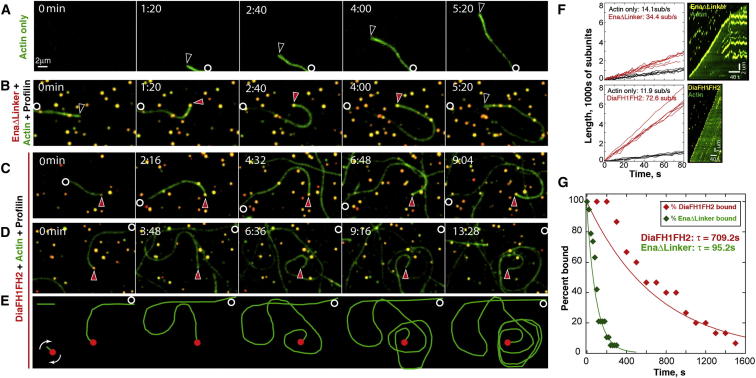
Figure 3.
Dia Is a Faster Elongator and Is More Processive than Ena
(A–D) TIRF montages, Movie S2: Oregon-green-labeled actin alone (A). Actin and Drosophila profilin with QD-biotin-SNAP-EnaΔLinker (B) or QD-biotin-SNAP-DiaFH1FH2 (C and D). Circles, filament pointed end; arrows, free filament barbed ends (open) or with EnaΔLinker or DiaFH1FH2 (red). QD blinks off in (D), but DiaFH1FH2 is present.
(E) Filament in (D) traced (green); QD (red).
(F) Filament elongation rates for controls (QD-free, black), EnaΔLinker (top left, red), or DiaFH1FH2 (bottom left, red). Representative kymographs (right) show single filaments with EnaΔLinker (top) or DiaFH1FH2 (bottom) processively bound to barbed end. Scale bars represent 2 μm (vertical) and 40 s (horizontal).
(G) Single exponential fit of percent bound versus time gives mean residence time (τ) for DiaFH1FH2 (red) and EnaΔLinker (green).