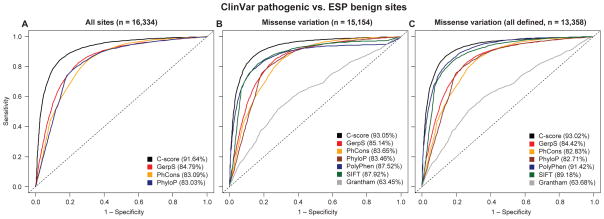
Figure 3.
Receiver operating characteristics (ROC) for discriminating curated, pathogenic mutations defined by the NIH ClinVar database27 matched to apparently benign ESP alleles (DAF ≥ 5%)24 with the same categorical consequence. The left panel shows genome-wide variants for which GerpS, PhCons, and PhyloP scores are defined (n=16,334), while the middle panel limits the analysis to missense changes (n=15,154), with missing values imputed to an upper value limit of each score, and right panel to missense changes for which PolyPhen, SIFT and Grantham scores are all defined (n=13,358). Versions of the right panel that exclude the overlap between PolyPhen training data and the ClinVar database or use a CADD model trained without PolyPhen as a feature are shown in Supplementary Fig. 12. Area under the curve (AUC) values are provided in the figure legend for each of the scores used.