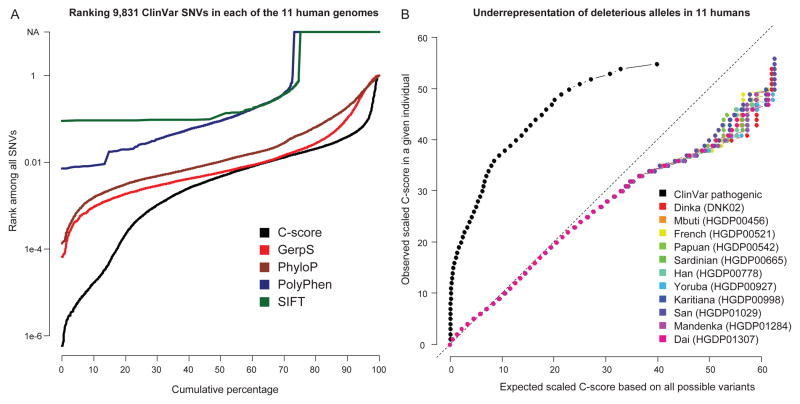
Figure 4.
Ranking of pathogenic ClinVar variants among the variants identified by whole genome sequencing of eleven human individuals from diverse populations. Left panel: Cumulative distributions of the ranks of 9,831 pathogenic ClinVar variants when “spiked in” to each of 11 personal genomes. For example, C-scores of ~30% of ClinVar variants rank in the top 0.1% of all variants within a personal genome, and most rank in the top 1%. About 25% of pathogenic ClinVar SNVs are not scored by PolyPhen/SIFT because of missing values or its restriction to missense variation; note also that ranks for PolyPhen/SIFT are computed among missense variants only and are therefore derived from far fewer total variants (see a plot restricted to missense variation in Supplementary Fig. 16). Right panel: A QQ-plot of the C-scores of the SNVs identified from the eleven individuals and pathogenic ClinVar SNVs. For a given scaled C-score observed in an individual, the fraction of that individual’s variants with a C-score at least that large was computed (y-axis). The C-score corresponding to this quantile of the distribution of all possible variants is displayed on the x-axis. High C-scores are underrepresented compared to the set of all possible variants. In contrast, known disease-causal variants from ClinVar have large C-scores relative to the set of all possible variants. This fact can be exploited to prioritize causal variants identified from whole genome sequencing of individual genomes (left panel and Supplementary Tables 10–11).