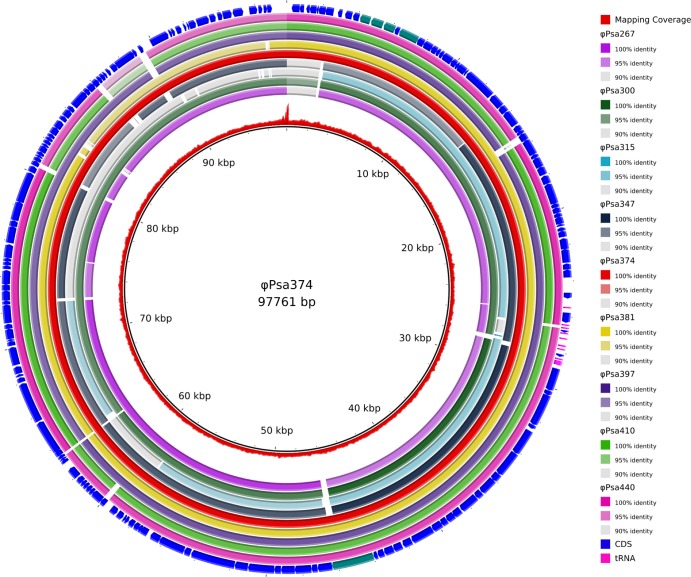
FIG 5.
Comparison of nine related Myoviridae genomes. The inner circle portrays the nonredundant reference genome of ϕPsa374, and the second inner-most ring shows the coverage of ϕPsa374 sequencing reads mapped to the assembled genome (shown as a red graph). The next nine colored rings (identified at right), representing the nine query phage genomes, indicate the sequence similarity (based on BLASTn) between ϕPsa374 and ϕPsa267, ϕPsa300, ϕPsa315, ϕPsa347, ϕPsa374 (included as a control), ϕPsa381, ϕPsa397, ϕPsa410, and ϕPsa440. Regions where the sequence similarity is less than 90% between the phages are represented as white gaps in the rings. The RAST-predicted CDS annotated in the ϕPsa374 genome are shown as arrows in the outer ring along with the predicted tRNA genes (in fuchsia). CDS predicted to be associated with tellurite resistance and the tape measure protein, as mentioned in the text, are highlighted in teal. The figure was prepared using the BLAST Ring Image Generator (BRIG) (60).