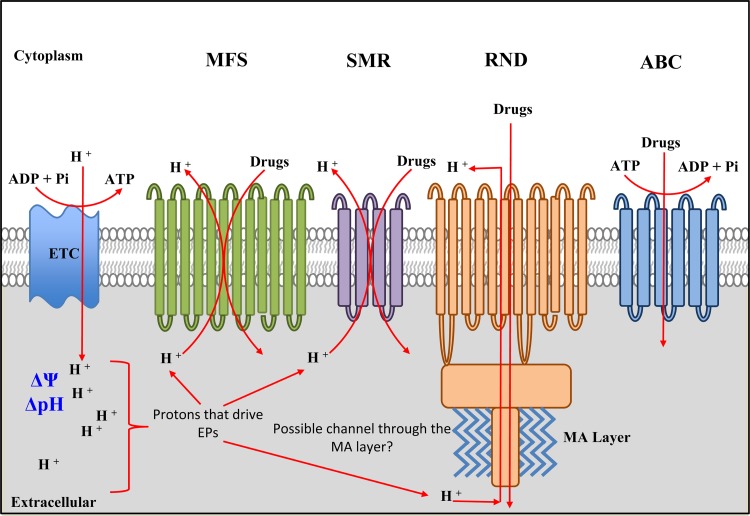
FIG 2.
Efflux pumps in M. tuberculosis. The EPs dependent on the PMF and ATP are shown. The mycobacterial ETC generates a transmembrane proton gradient (ΔpH) resulting in membrane potential (Δψ); ΔpH and Δψ together constitute the PMF. Protons translocated into the pseudoperiplasmic space are used by EPs to extrude drugs. The major facilitator superfamily (MFS) EPs are made of integral membrane proteins with 12 to 14 transmembrane regions, while the small multidrug resistance (SMR) EPs contain 4 to 6 transmembrane domains. Both systems use protons from the periplasm, and the members differ in their requirements for ΔpH and Δψ (see the text for details). The resistance nodulation/cell division (RND) EPs are also integral membrane proteins, and members of this family from Gram-negative bacteria associate with other proteins to form a multisubunit complex that spans both the inner and outer membranes. Hence, there is a possibility that a similar structure occurs in mycobacteria, where the members of this group of EPs span the pseudoperiplasmic space and associate with the mycolic acid (MA) layer, possibly through OmpA (outer membrane protein A)-like homologues. RND proteins also require protons—although these may originate from outside the cell—and the PMF. ABC transporters require ATP for the active drug extrusion and, as such, are also dependent on energy production in the cell. Not shown in the figure are the members of the fifth class of EPS, termed the multidrug and toxic compound extrusion (MATE) proteins; while some of these are dependent on PMF and protons, the majority of characterized members operate via sodium influx (93).