FIG 1.
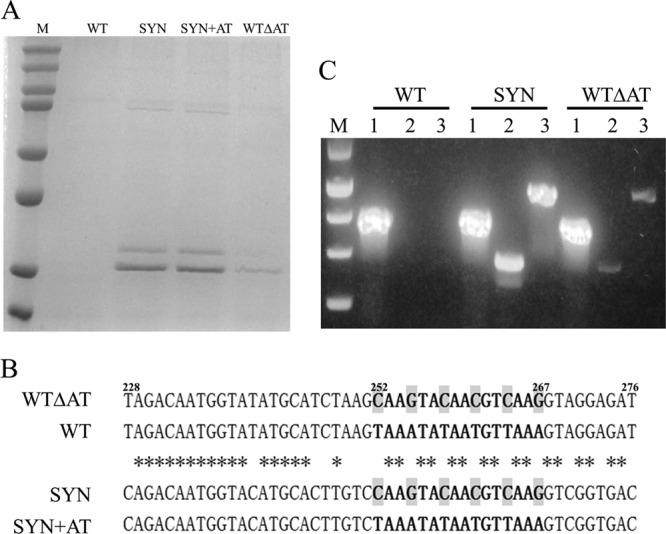
Gene sequence optimization enables high-level expression of LST in P. pastoris. (A) Comparison of LST expression levels from different recombinant genes. Lanes: M, protein marker (top to bottom, 250, 150, 100, 75, 50, 37, 25, and 20 kDa); WT, WT lst gene; SYN, SYN lst gene; SYN+AT, the A+T-rich region (nt 252 to 267) from the WT lst gene was introduced into the SYN lst gene; WTΔAT, the A+T-rich region (nt 252 to 267) of the WT lst gene was replaced by the corresponding sequence from the SYN lst gene. (B) Sequence alignment of a region from WT lst (nt 228 to 276) that caused the most significant decrease in expression when transplanted into the SYN lst gene. Asterisks indicate nucleotides that are identical among all four gene sequences. Shaded nucleotides are the six critical bases whose mutations were sufficient to activate LST expression. (C) Full-length LST mRNA and a shorter 5′ fragment were detected by RT-PCR. A fragment of AOX1 mRNA was used as an internal amplification and loading control. Lanes: M, DNA marker (top to bottom, 1,500, 1,000, 750, 500, and 250 bp); 1, fragment of AOX1; 2, 5′ fragment of LST mRNA; 3, full-length LST mRNA.
