Abstract
Significant food-borne disease outbreaks have occurred from consumption of ready-to-eat foods, including produce, contaminated with Listeria monocytogenes. Challenging food matrices (e.g., cantaloupe, sprouts) with limited processing steps postharvest to reduce pathogen loads have underscored a need for new mitigation strategies. Chlorine dioxide (ClO2) is increasingly being used in produce and other food systems to reduce food-borne pathogen levels. The goal of this study was to characterize the transcriptional response and survival of L. monocytogenes 10403S exposed to ClO2. The transcriptional profile of log-phase cells exposed to 300 mg/liter ClO2 for 15 min was defined by whole-genome microarray. A total of 340 genes were significantly differentially expressed. Among the differentially expressed genes, 223 were upregulated (fold change ≥ 1.5; adjusted P value < 0.05) in role categories responsible for protein fate, cellular processes, and energy metabolism. There were 113 and 16 genes differentially expressed belonging to regulatory networks of σB and CtsR, respectively. We assessed L. monocytogenes 10403S survival after exposure to 100, 300, and 500 mg/liter aqueous ClO2 in brain heart infusion (BHI) broth; there was a significant difference between cells exposed to 500 mg/liter ClO2 and those exposed to all other conditions over time (P value < 0.05). Isogenic ΔsigB and ΔctsR mutants exposed to 300 mg/liter ClO2 were more sensitive to ClO2 than the wild type under the same conditions. These results provide an initial insight into the mechanisms that L. monocytogenes employs to survive sublethal ClO2 and further our understanding of the inactivation mechanisms of this increasingly used sanitizer.
INTRODUCTION
Listeria monocytogenes is a Gram-positive, opportunistic food-borne pathogen which has the capacity to cause severe disease in humans and animals. Listeriosis can manifest as febrile gastroenteritis, sepsis, meningoencephalitis, and spontaneous abortion in pregnant women. There are approximately 1,600 cases of listeriosis in the United States annually, 99% of which are attributed to contaminated food (1). L. monocytogenes has been isolated from diverse environments (as reviewed in reference 2) and has the capacity to persist in agricultural (3), food manufacturing (4), and retail food (5) environments. Due to the high mortality risk of listeriosis (∼16% [1]), there is a definitive need for practical, safe, and effective mitigation strategies to control this organism from farm to fork. It is becoming more evident that fresh produce products transmit pathogens to humans (6). Next-generation sanitizers, such as chlorine dioxide (ClO2), are increasingly being used in the food industry, notably in produce (7). ClO2 is a strong, rapid oxidizing agent which is used in both gas and aqueous forms in food systems to reduce microbial loads. It effectively decreases microbial populations on the surfaces of foods (8) and food processing equipment (9). For example, ClO2 reduced L. monocytogenes levels by up to 4.3 CFU/cm2 on cantaloupe (10) and >3 log CFU/cm2 on stainless steel coupons (11). Evidence of degraded proteins extracted from Saccharomyces cerevisiae (12) and reduced commercially purified NADH to NAD+ in vitro (13) from ClO2 exposure has been proposed among the mechanisms of microbial inactivation. Subsequent studies in Escherichia coli have reported an observed drop in intracellular pH of surviving bacterial cells exposed to ClO2 (14) which results in cell death.
L. monocytogenes has many well-characterized stress response regulators that allow rapid, adaptive responses to intrinsic factors (e.g., salt and low pH) and to the intervention methods (e.g., high-pressure processing, low and high temperatures) used to control this pathogen in food. The roles of alternative sigma factors in regulation of genes related to stress response and virulence have been documented across many bacterial species as reviewed in Kazmierczak et al. (15). In L. monocytogenes, alternative sigma factors σB, σC, σH, and σL and other transcriptional regulators (e.g., CtsR and HrcA) play significant roles in stress response and resistance (16). Specifically, the general stress response sigma factor σB not only regulates genes that promote resistance to osmotic and acid stress but also increases the transcription of major virulence genes in L. monocytogenes (17). Exposure and adaptation to environmental stress result in cross-protection upon exposure to subsequent stresses. Evidence of cross-protection has been observed among stress conditions, including alkali, ethanol, and osmotic (18); heat and acid (19); and osmotic, oxidative, and low temperature (20) stresses. Exposure of L. monocytogenes to stress conditions (e.g., high acid, high salt) prior to ingestion by the host can lead to a more robust and efficient host cell invasion (21).
This study aimed to characterize transcriptional changes in L. monocytogenes exposed to ClO2 to better understand potential methods of inactivation and to elucidate stress response mechanisms that may facilitate adaptation, survival, and, potentially, tolerance. We hypothesized that L. monocytogenes employs multiple oxidative stress response systems as well as general stress response regulatory networks (e.g., σB) to overcome ClO2 oxidative stress. To test this hypothesis, we (i) identified changes in the global transcriptome after exposure to ClO2 and (ii) tracked the ability of wild-type L. monocytogenes 10403S and isogenic ΔsigB and ΔctsR mutants to survive ClO2 exposure over time. Our data suggest that several transcriptional regulatory networks, including σB and CtsR, and oxidative stress response systems help L. monocytogenes survive sublethal ClO2 exposure.
MATERIALS AND METHODS
Bacterial strains and storage.
L. monocytogenes 10403S (22) and its isogenic mutant strains L. monocytogenes 10403S ΔsigB (23) and L. monocytogenes 10403S ΔctsR (24) were used in this study. Stock cultures were stored at −80°C in brain heart infusion (BHI; Difco, Detroit, MI) broth–15% glycerol. Prior to each experiment, stock cultures were streaked onto BHI agar in order to obtain an isolated colony. A single colony was used to inoculate 5 ml of sterile BHI broth into a culture tube (16 by 125 mm). Following overnight incubation (15 to 18 h) at 37°C with 230 rpm shaking, 5 ml of preheated sterile BHI broth was inoculated with 50 μl of overnight culture. Following growth to the early log phase (optical density at 600 nm [OD600] = 0.4) at 37°C with 230 rpm shaking, 500 μl of early log phase was used to inoculate 50 ml of preheated sterile BHI broth into a 300-ml Nephelo flask (Bellco, Vineland, NJ). Cultures were grown to the early log phase (OD600 = 0.4) at 37°C and 230 rpm; these conditions were used for all phenotypic and transcriptional work.
Chlorine dioxide effect on log-phase cell survival.
Early log-phase cultures of L. monocytogenes 10403S were grown as described above and exposed to 100, 300, or 500 mg/liter commercially available aqueous chlorine dioxide (ClO2; CDG Environmental, Bethlehem, PA). A stock solution of ClO2 (3,000 mg/liter) was added directly to the broth culture at specific volumes to attain the target concentration. Prior to cell exposure, the ClO2 concentration of the stock solution was verified using the N,N-diethyl-p-phenylenediamine (DPD) method and the solution was scanned on a Hach DR/2500 spectrophotometer (Hach Instruments, Loveland, CO) (25). Phosphate-buffered saline (PBS) was added to the control samples at a volume equal to the volume of ClO2 added with the aim of minimizing the effects of volume and headspace differences on cell activity. Both PBS and ClO2 were preheated to 37°C prior to addition to the culture in order to limit the effects of temperature shifts on transcription. Cells exposed to 100, 300, or 500 mg/liter ClO2 were incubated at 37°C and 230 rpm, and a 500-μl aliquot was removed at 2.5, 5, 7.5, 10, 12.5, 15, and 20 min postexposure to enumerate cell populations. L. monocytogenes 10403S ΔsigB and ΔctsR were also exposed to 300 mg/liter ClO2 to assess survival as previously described. All exposed samples were combined with 500 μl of neutralizing buffer (BD Difco, Franklin Lakes, NJ) (1:1 ratio) to stop ClO2 activity and were stored on ice until further processing (≤30 min postexposure). All samples were serially diluted in PBS and spread plated onto BHI. Plates were incubated at 37°C; colonies were enumerated after 48 h of incubation. Statistical analyses were performed using SAS v 9.3 (SAS Institute, Inc., Cary, NC). A general linear model (GLM) and multivariate repeated measure analysis of variance (ANOVA) with a Tukey pairwise comparison were used to test the effects of “time” and “time*concentration” or “time*strain” interactions. Ratios (calculated as log CFU/ml treated − log CFU/ml untreated) were calculated to normalize each strain or condition to the initial cell population before ClO2 exposure. The variable “time” represented a repeated measure of the ratio of log CFU/ml cells treated with ClO2 to log CFU/ml unexposed cells after 2.5, 5, 7.5, 10, 12.5, 15, and 20 min. The variables “concentration” and “strain” represented different [ClO2] and isogenic deletion mutants. Significance was defined as P < 0.05. Three independent experiments were performed.
RNA extraction.
L. monocytogenes 10403S was grown to early log phase as previously described. Cells were exposed to 300 mg/liter aqueous ClO2 at 37°C and 230 rpm for 15 min. An equal volume of PBS was added to the control sample. Following exposure to ClO2, 50-ml cultures were combined with ice-cold neutralizing buffer (1:1 ratio) and immediately added to an ice-cold stop solution of 10% acid–phenol chloroform (Life Technologies, Grand Island, NY) in ethanol (10:1 ratio). Bacterial cells were collected by centrifugation at 15,300 × g for 20 min at 4°C. Cells were resuspended in 1 ml TRI reagent (Life Technologies, Grand Island, NY). Cell lysis was performed by bead beating (0.1 zircon beads) for 5 cycles of 30 s per cycle at speed setting 4 in a FastPrep FP120 device (BIO101/Savant, Santa Ana, CA). Samples were chilled on ice between cycles. Following centrifugation at 1,700 × g for 10 min at 4°C, 0.1 ml of 1-bromo-3-chloropropane was added to the supernatant and the mixture was incubated at room temperature for 10 min. RNA was precipitated in isopropanol at −20°C overnight. DNase treatment and further RNA purification were performed as outlined previously (26). RNA quantity and quality were assessed by spectroscopy using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE) and a 2100 Bioanalyzer system (Agilent, Foster City, CA), respectively. RNA integrity number (RIN) scores of greater than 7.0 were considered acceptable for further work.
cDNA synthesis and microarray hybridization.
Synthesis of cDNA from RNA was performed as previously outlined (27) with the exception of using SuperScript III reverse transcriptase (RT; Life Technologies, Grand Island, NY) for reverse transcription to cDNA. Quadruplicate samples were prepared with dye swapping to minimize dye bias.
The Cornell University Food Safety Laboratory (CUFSL) Listeria 6K oligonucleotide array (NCBI GEO GPL5029) was utilized as previously outlined (26, 28). This 70-mer oligonucleotide DNA microarray includes 2,857 open reading frames (ORFs) from L. monocytogenes EGD-e, 5 ORFs from Saccharomyces cerevisiae (negative control), L. monocytogenes chromosomal DNA (positive control), and salmon sperm DNA (printing quality control). It should be noted that the microarray used here was designed using the L. monocytogenes EGD-e genome sequence. The sequences of the targeted ORFs on the microarray were matched with the L. monocytogenes 10403S genome. Cross-hybridization index (CHI) values of 100%, >95%, and >90% were calculated for 2,107, 2,578, and 2,695 of the probes on the microarray, respectively (28). A total of 45 probes had a CHI value < 90, and 117 of the probes designed for the EGD-e genome were missing from the 10403S genome (28) and were omitted during analysis.
Preparation of the microarray slides, hybridization, and posthybridization washing and drying were performed as previously outlined (29). The microarrays were scanned using a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA) to provide working TIFF files.
Microarray statistical analysis.
Spots were aligned and identified by probe name within the software-guided grid and analyzed using GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA). Spots that were empty or saturated in both channels or had poor morphology were flagged and removed from further analysis. Microarray data were analyzed using LIMMA (linear models for microarray data) package version 3.18.2 (30) through the use of BioConductor software supported by R version 2.15.1 (31). Background correction (normexp method), within-array normalization (print-tip loess method), and between-array normalization (scale method) were all used for data preprocessing (32, 33). Differential expression analysis was performed by first fitting a linear model to the normalized data followed by a smoothing correction (eBays function) (30). Differentially expressed genes were defined as having a fold change (FC) ≥ 1.50 and an adjusted P value < 0.05. The activity of defined operons was based on those previously annotated by our group through transcriptome sequencing (RNA-Seq) analyses (34). The continuity of RNA-Seq coverage across neighboring genes and the presence or absence of Rho-independent terminators were used for operon characterization (34). Differentially expressed genes were categorized into the published J. Craig Venter Institute (JCVI) role categories and functional groups (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi) as utilized previously to investigate regulatory networks (17). An overall χ2 test was performed to determine if role categories were over- and underrepresented; odds ratios (OD) were calculated to identify which categories were over- and underrepresented. The Department of Energy Joint Genome Institute Integrated Microbial Genomes (DOE JGI IMG) (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) listings were consulted further for additional functional group annotation.
TaqMan qRT-PCR.
TaqMan primers (IDT DNA, Coralville, IA) and probes (Applied Biosystems, Foster City, CA) were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA) for L. monocytogenes 10403S genes rpoB, lexA, clpC, dnaK, sigB, lmo0669, and lmo1433 (Table 1). Quantitative reverse transcriptase PCR (qRT-PCR) was performed as outlined (35) with TaqMan One-Step RT-PCR Master Mix reagent and MultiScribe RT for single-tube reverse transcription and amplification using a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). Negative controls to detect genomic DNA contamination were performed by omitting reverse transcriptase for each replicate and primer and probe set. All reactions were performed in duplicate for each of three independent RNA isolations. A standard curve based on genomic DNA copy number was generated for each gene. Log-transformed (log10) values of mRNA expression values were normalized against the expression value of the housekeeping gene rpoB, which is routinely used for normalization of qRT-PCR data in L. monocytogenes (35). Statistical analyses were performed using SAS v 9.3 (SAS Institute, Inc., Cary, NC) with a general linear model (GLM) and Dunnett correction for multiple comparisons. Significantly different transcript levels between the experimental and control samples were defined by a P value < 0.05.
TABLE 1.
TaqMan primers and probesa
| Target | Forward primer | Probeb | Reverse primer |
|---|---|---|---|
| rpoB | CGATCTTGGAGAGCCGAAATA | CGGTAGAAGAATCTAAGAAC | GAGCCGCATAGTTTGCATCAC |
| sigB | GTAGAGTCCATCGCCCGAAA | TTCCCAAGGTAAATCT | TCCCAACTTGAACTAAATCTTCATGA |
| lmo0669 | TGAAAAGCAACGCCAGGATT | CCTGGTTTGCAAAGTA | TCGCCCGCATCTGGAA |
| lmo1433 | AGCACAGGCAGCAGGGTTAA | AGCAATTGTAGAAGAGCG | GTACACACGTTCCACCCCAAT |
| clpC | AAAGCTCGGCAGCAAGTTCTA | TTAGGCGGCGGTGAT | GTCTCCCCGCCCCAGTAG |
| dnaK | CGAACGTAATACAACTATCCCAACA | AAATCACAAACTTTCTCTACAGC | ATCTACGGCTGGTTGGTTATCAG |
| lexA | ACTGTTCATGGGCATCTTGCT | CTTGAAGGTAAAGGGC | GAGGCTTTGTAGGGTCACGTCTA |
All sequences are written 5′ → 3′.
All probes have a 5′ 6-carboxyfluorescein (FAM) reporter dye with a 3′ MGB dark quencher dye.
Microarray data accession number.
The raw and normalized microarray data are available through the NCBI Gene Expression Omnibus (GEO) genomics data repository under accession number GSE48187.
RESULTS
Effect of [ClO2] on L. monocytogenes 10403S survival.
Early log-phase cultures of L. monocytogenes 10403S were exposed to three concentrations of aqueous ClO2 (Table 2) to (i) assess survival and (ii) determine exposure concentrations that would elicit a transcriptional effect for subsequent microarray analyses. The variables of “time,” “concentration,” and “time*concentration” had a significant effect on L. monocytogenes survival (P values = 0.0003, 0.0084, and 0.0001, respectively). There was a significant difference in survival between cells exposed to 500 mg/liter ClO2 and those exposed to 300 and 100 mg/liter after 12.5 min (Table 2). Specifically, fewer cells survived the 500 mg/liter ClO2 treatment. Exposure of cells to 300 mg/liter ClO2 for 15 min resulted in a decrease in cell population that was not statistically significant; 300 mg/liter ClO2 was selected as a suitable condition to investigate transcriptional effects of sublethal ClO2 concentrations due to the observable subtle effect on cell populations.
TABLE 2.
L. monocytogenes ClO2 stress survival over time
| [ClO2] or strain | Ratio ± SD of log CFU/ml cells treated with ClO2 to log CFU/ml unexposed cells at indicated time (min)b |
||||||
|---|---|---|---|---|---|---|---|
| 2.5 | 5 | 7.5 | 10 | 12.5 | 15 | 20 | |
| [ClO2] (mg/liter)a | |||||||
| 100 | −0.14 ± 0.30A | −0.31 ± 0.19A | −0.34 ± 0.19A | −0.34 ± 0.16A | −0.34 ± 0.13A | −0.25 ± 0.11A | 0.20 ± 0.18A |
| 300 | 0.02 ± 0.26A | −0.13 ± 0.38A | −0.06 ± 0.17A | −0.06 ± 0.10A | −0.17 ± 0.05A | −0.33 ± 0.37A | −0.21 ± 0.15A |
| 500 | −0.90 ± 0.62A | −1.45 ± 1.51A | −1.65 ± 1.45A | −1.97 ± 1.42A | −2.59 ± 1.07B | −3.65 ± 0.55B | −3.65 ± 0.55B |
| Strain | |||||||
| Wild typec | 0.02 ± 0.26A | −0.13 ± 0.38A | −0.06 ± 0.17A | −0.06 ± 0.10A | −0.17 ± 0.05A | −0.33 ± 0.37A | −0.21 ± 0.15A |
| ΔsigBd | −0.59 ± 0.17B | −0.61 ± 0.10A | −0.59 ± 0.18B | −0.59 ± 0.14C | −0.69 ± 0.14A | −0.65 ± 0.21B | −0.67 ± 0.21B |
| ΔctsRe | −0.37 ± 0.16B | −0.38 ± 0.34A | −0.42 ± 0.26B | −0.42 ± 0.12B | −0.44 ± 0.38A | −0.40 ± 0.32AB | −0.42 ± 0.18A |
Concentration of ClO2 added to log-phase wild-type L. monocytogenes 10403S.
Different superscript letters denote statistically significant ratios among concentrations or strains at that time point.
Ratio of log CFU/ml wild-type cells treated with 300 mg/liter ClO2 to log CFU/ml unexposed wild-type cells after 2.5, 5, 7.5, 10, 12.5, 15, and 20 min.
Ratio of log CFU/ml ΔsigB cells treated with ClO2 to log CFU/ml unexposed ΔsigB cells after 2.5, 5, 7.5, 10, 12.5, 15, and 20 min.
Ratio of log CFU/ml ΔctsR cells treated with ClO2 to log CFU/ml unexposed ΔctsR cells after 2.5, 5, 7.5, 10, 12.5, 15, and 20 min.
Microarray analyses identified 340 differentially expressed genes upon exposure to ClO2.
Whole-genome microarray analyses identified 340 genes as significantly differentially expressed following exposure of log-phase L. monocytogenes 10403S to ClO2. These genes constituted 114 putative operons among which at least one gene showed differential expression. Of the 340 genes differentially expressed, 223 genes were upregulated (≥1.5-fold change [FC]; adjusted P value < 0.05) upon exposure to ClO2 compared to unexposed wild-type cells (Table 3; see also Table S1 in the supplemental material). In contrast, 117 genes were significantly downregulated in cells exposed to ClO2 compared to unexposed cells (FC ≥ 1.50; adjusted P value < 0.05) (Table 4; see also Table S2).
TABLE 3.
Genes upregulated upon exposure to ClO2 among over- and underrepresented JCVI role categoriesa
| Locus tagb | Gene name | Annotationc | Fold change | Adjusted P value |
|---|---|---|---|---|
| Cell envelope | ||||
| lmo0584 | Conserved hypothetical membrane protein | 2.58 | 0.006 | |
| lmo0880 | Similar to wall-associated protein precursor (LPXTG motif) | 8.68 | 0.005 | |
| Cellular processes | ||||
| lmo0220 | ftsH | Highly similar to cell division protein FtsH | 1.55 | 0.005 |
| lmo0321 | Similar to unknown proteins | 4.86 | 0.003 | |
| lmo0515 | Conserved hypothetical protein | 8.05 | 0.018 | |
| lmo0669 | Similar to oxidoreductase | 5.78 | 0.049 | |
| lmo0906 | Similar to glutathione reductase | 1.84 | 0.038 | |
| lmo0983 | Similar to glutathione peroxidase | 2.76 | 4E-04 | |
| lmo1288 | Conserved hypothetical protein | 1.9 | 0.01 | |
| lmo1433 | Similar to glutathione reductase | 2.53 | 0.021 | |
| lmo1439 | sod | Superoxide dismutase | 2.44 | 2E-04 |
| lmo1577 | Similar to unknown proteins | 2.07 | 0.012 | |
| lmo1580 | Similar to unknown protein | 4.76 | 0.002 | |
| lmo1601 | Similar to general stress protein | 3.37 | 0.003 | |
| lmo1694 | Similar to CDP-abequose synthase | 2.64 | 0.014 | |
| lmo1708 | Similar to aminoglycoside N3-acetyltransferases | 1.57 | 0.03 | |
| lmo1879 | cspD | Similar to cold shock protein | 3.87 | 0.004 |
| lmo2016 | cspB | Similar to major cold shock protein | 1.76 | 0.002 |
| lmo2190 | mecA | Competence negative regulator MecA | 1.87 | 0.002 |
| lmo2230 | Similar to arsenate reductase | 8.07 | 1E-03 | |
| lmo2398 | ltrC | Low-temperature-requirement C protein, also similar to B. subtilis YutG protein | 2.39 | 0.005 |
| lmo2426 | Conserved hypothetical proteins | 1.81 | 0.003 | |
| lmo2673 | Conserved hypothetical protein | 8.85 | 0.003 | |
| Energy metabolism | ||||
| lmo0232 | clpC | Endopeptidase Clp ATP-binding chain C | 4.84 | 3E-04 |
| lmo0261 | Similar to phospho-beta-glucosidase | 1.98 | 0.03 | |
| lmo0268 | Similar to phosphoglycerate mutase | 6.77 | 1E-04 | |
| lmo0342 | Similar to transketolase | 2.96 | 0.014 | |
| lmo0347 | Similar to dihydroxyacetone kinase | 4.54 | 0.024 | |
| lmo0521 | Similar to 6-phospho-beta-glucosidase | 1.82 | 0.049 | |
| lmo0613 | Similar to oxidoreductase | 5.22 | 2E-04 | |
| lmo0646 | Similar to unknown proteins | 4.18 | 2E-04 | |
| lmo0722 | Similar to pyruvate oxidase | 2.96 | 0.007 | |
| lmo0758 | Unknown | 118 | 2E-07 | |
| lmo0759 | Unknown | 141 | 2E-07 | |
| lmo0781 | Similar to mannose-specific phosphotransferase system (PTS) component IID | 6.35 | 6E-04 | |
| lmo0811 | Similar to carbonic anhydrase | 2.2 | 0.015 | |
| lmo0907 | Similar to phosphoglycerate mutase | 2.27 | 0.008 | |
| lmo0913 | Similar to succinate semialdehyde dehydrogenase | 8.15 | 0.002 | |
| lmo0915 | Similar to phosphotransferase system enzyme IIC | 12.5 | 0.009 | |
| lmo0917 | 11.5 | 0.042 | ||
| lmo0943 | fri | Nonheme iron-binding ferritin | 3.23 | 0.007 |
| lmo1054 | 1.48 | 0.046 | ||
| lmo1055 | pdhD | Highly similar to dihydrolipoamide dehydrogenase, E3 subunit of pyruvate dehydrogenase complex | 1.98 | 0.006 |
| lmo1233 | trxA | Thioredoxin | 2.37 | 0.023 |
| lmo1254 | Similar to alpha,alpha-phosphotrehalase | 7.47 | 2E-04 | |
| lmo1255 | Similar to PTS system trehalose-specific enzyme IIBC | 6.61 | 2E-04 | |
| lmo1293 | glpD | Similar to glycerol-3-phosphate dehydrogenase | 3.11 | 0.009 |
| lmo1348 | Similar to aminomethyltransferase | 5.98 | 0.022 | |
| lmo1350 | Similar to glycine dehydrogenase (decarboxylating) subunit 2 | 4.84 | 0.034 | |
| lmo1514 | Similar to unknown protein | 1.74 | 0.007 | |
| lmo1538 | Similar to glycerol kinase | 11.9 | 5E-05 | |
| lmo1579 | Similar to alanine dehydrogenase | 2.34 | 0.006 | |
| lmo1789 | Weakly similar to NAD(P)H oxidoreductase chain B | 1.62 | 0.041 | |
| lmo1883 | Similar to chitinases | 3.17 | 0.031 | |
| lmo1995 | dra | Similar to deoxyribose-phosphate aldolase | 1.91 | 0.001 |
| lmo2000 | Similar to PTS mannose-specific enzyme IID component | 2.47 | 0.039 | |
| lmo2057 | ctaB | Highly similar to heme A farnesyltransferase | 1.85 | 0.015 |
| lmo2122 | Similar to maltodextrose utilization protein MalA | 4.29 | 0.017 | |
| lmo2205 | Similar to phosphoglyceromutase 1 | 2.2 | 0.038 | |
| lmo2363 | Similar to glutamate decarboxylase | 1.78 | 0.031 | |
| lmo2373 | Similar to phosphotransferase system (PTS) beta-glucoside-specific enzyme IIB component | 1.98 | 0.036 | |
| lmo2425 | 1.45 | 0.01 | ||
| lmo2437 | Unknown | 1.6 | 0.027 | |
| lmo2494 | Similar to negative regulator of phosphate regulon | 2.37 | 0.012 | |
| lmo2528 | atpC | Highly similar to H+-transporting ATP synthase chain epsilon | 1.77 | 0.006 |
| lmo2573 | Similar to zinc-binding dehydrogenase | 1.98 | 0.038 | |
| lmo2650 | Similar to hypothetical PTS enzyme IIB component | 5.69 | 0.002 | |
| lmo2674 | Similar to ribose-5-phosphate epimerase | 5.26 | 0.002 | |
| lmo2724 | Similar to unknown proteins | 2.44 | 0.004 | |
| lmo2743 | Similar to transaldolase | 2.31 | 0.01 | |
| lmo2830 | Similar to thioredoxin | 1.89 | 0.012 | |
| Hypothetical protein | ||||
| lmo0170 | Unknown | 2.72 | 0.003 | |
| lmo0229 | ctsR | Highly similar to transcription repressor of class III stress genes (CtsR) | 4.82 | 0.042 |
| lmo0231 | Similar to arginine kinase | 2.2 | 0.043 | |
| lmo0439 | Weakly similar to a module of peptide synthetase | 2.44 | 0.02 | |
| lmo0496 | Similar to B. subtilis YnzC protein | 4.93 | 7E-04 | |
| lmo0596 | Similar to unknown proteins | 4.09 | 0.002 | |
| lmo0670 | Unknown | 4.49 | 1E-03 | |
| lmo0720 | Unknown | 2.3 | 0.041 | |
| lmo0761 | Similar to unknown proteins | 36.8 | 4E-05 | |
| lmo0796 | Conserved hypothetical protein | 15.9 | 0.001 | |
| lmo0869 | Unknown | 2.9 | 0.035 | |
| lmo0911 | Unknown | 2.58 | 0.023 | |
| lmo0964 | Similar to B. subtilis YjbH protein | 2.28 | 0.022 | |
| lmo1059 | Unknown | 1.79 | 0.003 | |
| lmo1069 | Similar to B. subtilis YlaI protein | 1.68 | 0.008 | |
| lmo1137 | Unknown | 2.65 | 2E-04 | |
| lmo1332 | Similar to conserved hypothetical proteins | 1.53 | 0.038 | |
| lmo1501 | Similar to unknown proteins | 1.55 | 0.023 | |
| lmo1502 | Similar to unknown proteins | 1.72 | 0.013 | |
| lmo1503 | Unknown | 1.52 | 0.019 | |
| lmo1526 | Similar to unknown proteins | 4.24 | 0.002 | |
| lmo1535 | Similar to unknown proteins | 1.88 | 0.023 | |
| lmo1602 | Similar to unknown proteins | 3.15 | 0.007 | |
| lmo1608 | Similar to unknown proteins | 2.54 | 0.001 | |
| lmo1704 | Similar to conserved hypothetical proteins | 3.67 | 0.001 | |
| lmo1728 | Some similarities to cellobiose-phosphorylase | 4.33 | 0.021 | |
| lmo1921 | 1.47 | 0.027 | ||
| lmo2054 | Similar to unknown proteins | 1.55 | 0.014 | |
| lmo2191 | spxA | Similar to unknown proteins | 2.12 | 0.006 |
| lmo2213 | Similar to unknown protein | 23.6 | 5E-05 | |
| lmo2387 | Conserved hypothetical protein | 2.39 | 0.025 | |
| lmo2391 | Conserved hypothetical protein similar to B. subtilis YhfK protein | 10.8 | 2E-04 | |
| lmo2392 | 1.46 | 0.023 | ||
| lmo2393 | 1.47 | 0.03 | ||
| lmo2570 | Unknown | 3.18 | 0.003 | |
| lmo2572 | Similar to chain A, dihydrofolate reductase | 3.6 | 5E-04 | |
| lmo2585 | Similar to B. subtilis YrhD protein | 16.1 | 0.003 | |
| lmo2603 | Unknown | 2.3 | 0.014 | |
| lmo2646 | Unknown | 6.42 | 1E-03 | |
| lmo2692 | Unknown | 1.52 | 0.03 | |
| lmo2705 | Unknown | 1.52 | 0.031 | |
| Protein fate | ||||
| lmo0764 | Similar to lipoate-protein ligase | 4.72 | 0.001 | |
| lmo1051 | Similar to formylmethionine deformylase and to B. subtilis YkrB protein | 1.55 | 0.024 | |
| lmo1268 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | 1.79 | 0.018 |
| lmo1354 | Similar to aminopeptidase P | 1.68 | 0.026 | |
| lmo1473 | dnaK | class I heat shock protein (molecular chaperone) DnaK | 2.02 | 0.024 |
| lmo1474 | grpE | Heat shock protein GrpE | 2.22 | 0.008 |
| lmo1578 | Similar to X-Pro dipeptidase | 2.44 | 0.004 | |
| lmo1611 | Similar to aminopeptidase | 1.97 | 0.004 | |
| lmo1620 | 1.5 | 0.031 | ||
| lmo2068 | groEL | Class I heat shock protein (chaperonin) GroEL | 2.22 | 7E-04 |
| lmo2069 | groES | Class I heat shock protein (chaperonin) GroES | 2.51 | 0.002 |
| lmo2199 | ohrA | Similar to unknown protein | 9.67 | 3E-06 |
| lmo2206 | clpB | Similar to endopeptidase Clp ATP-binding chain B (ClpB) | 3.07 | 0.012 |
| lmo2398 | ltrC | Low-temperature-requirement C protein, also similar to B. subtilis YutG protein | 2.39 | 0.005 |
| lmo2468 | clpP | ATP-dependent Clp protease proteolytic subunit | 2.54 | 0.009 |
| lmo2510 | 1.5 | 0.031 | ||
| Protein synthesis | ||||
| lmo0211 | ctc | Similar to B. subtilis general stress protein | 2.58 | 0.02 |
| lmo0240 | Highly similar to B. subtilis YazC protein | 2.03 | 0.004 | |
| lmo1218 | Similar to rRNA methylase | 1.68 | 0.013 | |
| lmo1703 | Similar to RNA methyltransferases | 1.52 | 0.039 | |
| lmo2511 | Similar to conserved hypothetical proteins such as B. subtilis YvyD protein | 6.26 | 0.003 |
All genes that showed a higher transcript level in the ClO2-exposed samples over the control sample without treatment are listed with an adjusted P value < 0.05 and fold change of ≥1.50. Only genes categorized into JCVI role categories with over- and underrepresentation on the basis of the odds ratio are listed.
Locus tag identification is on the basis of the lmo number from the L. monocytogenes EGD-e strain.
Annotation was defined on the basis of the JCVI role category listings.
TABLE 4.
Genes downregulated upon exposure to ClO2 among over- and underrepresented JCVI role categoriesa
| Locus tagb | Gene name | Annotationc | Fold change | Adjusted P value |
|---|---|---|---|---|
| Cell envelope | ||||
| lmo0198 | gcaD | Highly similar to UDP-N-acetylglucosamine pyrophosphorylase | 1.604 | 0 |
| lmo0695 | Unknown | 1.88 | 0.01 | |
| lmo0971 | dltD | DltD protein for d-alanine esterification of lipoteichoic acid and wall teichoic acid | 1.959 | 0.05 |
| lmo0972 | dltC | d-Alanyl carrier protein | 2.081 | 0 |
| lmo0973 | dltB | DltB protein for d-alanine esterification of lipoteichoic acid and wall teichoic acid | 2.265 | 0 |
| lmo1082 | Similar to dTDP-sugar epimerase | 1.749 | 0.01 | |
| lmo1548 | mreB | Similar to cell-shape determining protein MreB | 1.977 | 0 |
| lmo2038 | murE | Similar to UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | 1.793 | 0.04 |
| lmo2549 | gtcA | Wall teichoic acid glycosylation protein GtcA | 1.774 | 0.02 |
| Cellular processes | ||||
| lmo0217 | Similar to B. subtilis DivIC protein | 1.955 | 0.01 | |
| lmo0677 | Similar to flagellar biosynthesis protein FliQ | 4.934 | 0 | |
| lmo0679 | Similar to flagellar biosynthetic protein FlhB | 1.934 | 0.02 | |
| lmo0693 | Similar to flagellar motor switch protein FliY C-terminal part | 1.506 | 0.04 | |
| lmo0696 | Similar to flagellar hook assembly protein | 1.731 | 0.02 | |
| lmo1071 | Similar to cell division protein RodA and FtsW | 1.885 | 0.01 | |
| lmo1297 | Similar to aluminum resistance protein and to B. subtilis YnbB protein (hypothetical) | 1.604 | 0.05 | |
| lmo1364 | cspL | Similar to cold shock protein | 1.808 | 0 |
| lmo1544 | minD | Highly similar to cell division inhibitor (septum placement) protein MinD | 1.594 | 0.02 |
| lmo1699 | Some similarities to methyl-accepting chemotaxis proteins | 2.075 | 0 | |
| lmo1700 | Unknown | 1.782 | 0.03 | |
| lmo2040 | ftsL | Similar to cell division protein FtsL | 1.738 | 0.02 |
| lmo2427 | Similar to cell division proteins RodA, FtsW | 1.73 | 0.05 | |
| lmo2428 | Similar to cell division proteins RodA, FtsW | 1.996 | 0.01 | |
| lmo2506 | ftsX | Highly similar to cell division protein FtsX | 1.81 | 0.03 |
| lmo2507 | ftsE | Highly similar to the cell division ATP-binding protein FtsE | 1.828 | 0.01 |
| lmo2569 | Similar to dipeptide ABC transporter (dipeptide-binding protein) | 1.635 | 0.04 | |
| Energy metabolism | ||||
| lmo1072 | pycA | Highly similar to pyruvate carboxylase | 1.521 | 0.04 |
| lmo2367 | pgi | Glucose-6-phosphate isomerase | 1.597 | 0.01 |
| Fatty acid and phospholipid metabolism | ||||
| lmo0970 | Similar to enoyl-acyl-carrier protein reductase | 1.634 | 0.02 | |
| lmo1806 | acpA | Highly similar to acyl carrier proteins | 2.068 | 0.01 |
| lmo1807 | fabG | Similar to 3-ketoacyl-acyl-carrier protein reductase | 1.828 | 0.01 |
| lmo2202 | Similar to 3-oxoacyl-acyl-carrier protein synthase | 2.412 | 0 | |
| lmo2450 | Similar to carboxylesterase | 1.803 | 0.02 | |
| Protein synthesis | ||||
| lmo0177 | metS | Methionyl-tRNA synthetase | 1.811 | 0 |
| lmo0244 | Similar to ribosomal protein L33 type II | 1.944 | 0 | |
| lmo0695 | Unknown | 1.88 | 0.01 | |
| lmo1294 | miaA | Similar to tRNA isopentenylpyrophosphate transferase | 2.196 | 0.01 |
| lmo1658 | rpsB | 30S ribosomal protein S2 | 1.55 | 0.03 |
| lmo1755 | gatA | Glutamyl-tRNA (Gln) amidotransferase (subunit A) | 1.506 | 0.02 |
| lmo2047 | rpmF | Ribosomal protein L32 | 2.195 | 0 |
| lmo2597 | rplM | Ribosomal protein L13 | 2.077 | 0 |
| lmo2633 | rpsJ | Ribosomal protein S10 | 1.523 | 0.02 |
| lmo2811 | Similar to GTPase | 1.727 | 0.02 | |
| lmo2856 | rpmH | Ribosomal protein L34 | 1.979 | 0.02 |
| Purines, pyrimidines, nucleosides, and nucleotides | ||||
| lmo1827 | Similar to guanylate kinases | 1.771 | 0 | |
| lmo1929 | ndk | Similar to nucleoside diphosphate kinase | 2.104 | 0 |
| lmo0509 | prs | Similar to phosphoribosyl pyrophosphate synthetase | 1.768 | 0.01 |
| lmo1096 | guaA | Highly similar to GMP synthetase | 2.904 | 0 |
| lmo1929 | ndk | Similar to nucleoside diphosphate kinase | 2.104 | 0 |
| lmo1463 | Similar to cytidine deaminase | 1.75 | 0.01 | |
| lmo1885 | Similar to xanthine phosphoribosyltransferase | 2.201 | 0.01 | |
| Unknown function | ||||
| lmo1240 | Conserved hypothetical protein, similar to B. subtilis YsnB protein | 1.763 | 0 | |
| lmo0998 | Similar to hypothetical protein | 1.914 | 0.02 | |
| lmo1067 | Similar to GTP-binding elongation factor | 2.936 | 0 | |
| lmo1479 | lepA | Highly similar to GTP-binding protein LepA | 1.544 | 0.01 |
| lmo1500 | Similar to unknown proteins | 2.351 | 0.02 | |
| lmo2779 | Similar to probable GTP-binding protein | 2.916 | 0 | |
| lmo2802 | gidB | GidB protein | 1.511 | 0.03 |
All genes that showed a lower transcript level in the ClO2-exposed samples over the control sample without treatment are listed with an adjusted P value < 0.05 and a fold-change of ≥1.50. Only genes categorized into JCVI role categories with over- and underrepresentation on the basis of the odds ratio are listed.
Locus tag identification is on the basis of the lmo number from the L. monocytogenes EGD-e strain.
Annotation was defined on the basis of the JCVI role category listings.
Six JCVI role categories were either over- or underrepresented among significantly upregulated genes.
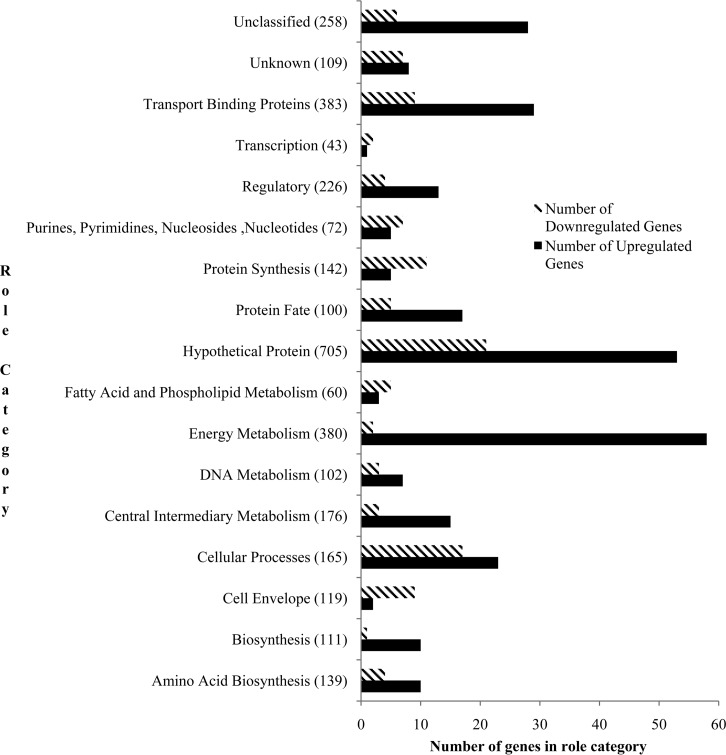
Genes that were upregulated in cells exposed to ClO2 were classified by their JCVI role categories (Fig. 1) to identify categories that were over- and underrepresented after stress exposure (Table 5). The energy metabolism role category was overrepresented (OR = 2.1) and included six genes encoding components of phosphotransferase systems (PTS), lmo0613 (oxidoreductase), lmo2830 (thioredoxin), lmo1789 [NAD(P)H oxidoreductase chain B], and lmo2528 (proton-transporting ATP synthase chain). The cellular processes role category group was overrepresented (OR = 1.7) and included genes lmo0669 (oxidoreductase), lmo0906 and lmo1433 (glutathione reductases), and lmo1439 (superoxide dismutase). The activity of genes encoding enzymes such as oxidoreductase or glutathione reductase would allow increased resistance or adaptability to oxidative stress. The protein fate role category was overrepresented as well (OR = 2.2); seven of the genes in that category encoded heat shock proteins. A large representation of transcriptional activity of genes encoding chaperone proteins suggests a demand for protein folding and degradation. As mentioned, ctsR, the regulator for class III heat shock genes, was upregulated. In addition, hrcA, the regulator for class I heat shock genes, was upregulated but not found to be statistically significantly differentially expressed (FC = 1.7; adjusted P value = 0.05). The protein synthesis (OR = 0.4) and cell envelope (OR = 0.2) role categories were underrepresented.
FIG 1.
Differentially expressed gene distribution among JCVI role categories. The JCVI role categories are listed on the y axis. The total number of genes in each role category is in parentheses. Black bars represent the numbers of genes upregulated following ClO2 exposure; hashed bars represent the numbers of genes downregulated following exposure to ClO2.
TABLE 5.
JCVI role categories over- or underrepresented for genes upregulated upon exposure of strains to ClO2
| JCVI role categorya | No. of annotated genes | No. of positively expressed genes in ClO2-exposed strainsb | Odds ratioc | Chi-square test P valued |
|---|---|---|---|---|
| Cellular processes | 165 | 21 | 1.8 | 0.018 |
| Energy metabolism | 380 | 48 | 1.8 | 0.003 |
| Protein fate | 100 | 16 | 2.3 | 0.002 |
| Protein synthesis | 142 | 5 | 0.4 | 0.048 |
| Cell envelope | 119 | 2 | 0.2 | 0.010 |
| Hypothetical | 705 | 41 | 0.7 | 0.021 |
JCVI role categories for L. monocytogenes EDG-e.
Number of genes upregulated upon ClO2 exposure within the selected role category.
Odd ratios were employed to characterize the representation within each category.
The chi-square test was used to identify the significant association between the number of genes overly active and the number of total genes within the selected role category. A P value of <0.05 was used to determine significance.
Significantly downregulated genes were either over- or underrepresented in 7 JCVI role categories.
Genes that were downregulated in cells exposed to ClO2 were categorized by their JCVI role categories (Fig. 1) to identify categories that were over- or underrepresented after stress exposure (Table 6). The cell envelope role category was overrepresented (OR = 2.3); this group includes genes responsible for cell wall manufacturing, specifically, teichoic acid formation and accumulation (gtcA, dltD, and dltC) and cell shape determination (mreB). Similarly to the upregulated genes, the cell processes category was also overrepresented among downregulated genes (OR = 3.5). The genes downregulated in this role category include lmo0217 (similar to divIC septum formation), minD (septum placement), lmo1071, lmo2427, and lmo2428 (similar to genes encoding cell division proteins RodA and FtsW), and ftsL, ftsX, and ftsE (cell-division proteins). It should be noted that there was increased expression of minC (FC = 1.47; adjusted P value = 0.028), but constraints were not met for minC to be considered significantly differentially expressed. MinC is responsible for septum site determination. The fatty acid and phospholipid metabolism category was overrepresented (OR = 2.6) and included genes encoding acyl-carrier proteins (lmo0970, lmo1806, lmo1807, lmo2202, and lmo2450) responsible for membrane lipid production (36), which suggests reduced availability of membrane structural components. This correlates well with decreased transcription of genes encoding teichoic acid production. The purines, pyrimidines, nucleosides, and nucleotides role category, which includes ndk, prs, guaA, and lmo1827, was also overrepresented (OR = 3.1). Although not included in this role category, lepA, encoding a GTP-binding protein that inhibits tRNA through ribosomal back-translocase activity, was also downregulated. The energy metabolism role category was underrepresented among downregulated genes, but it was overrepresented by upregulated genes.
TABLE 6.
JCVI role categories over- or underrepresented for genes downregulated upon exposure of strains to ClO2
| JCVI role categorya | No. of annotated genes | No. of positively expressed genes in ClO2-exposed strainsb | Odds ratioc | Chi-square test P valued |
|---|---|---|---|---|
| Cell envelope | 119 | 9 | 2.3 | 0.015 |
| Cellular processes | 165 | 17 | 3.5 | <0.001 |
| Fatty acid and phospholipid metabolism | 60 | 5 | 2.6 | 0.042 |
| Protein synthesis | 142 | 11 | 2.4 | 0.005 |
| Purines, pyrimidines, nucleosides, nucleotides | 72 | 7 | 3.1 | 0.004 |
| Unknown | 109 | 7 | 2.2 | 0.030 |
| Energy metabolism | 380 | 2 | 0.13 | 0.001 |
JCVI role categories for L. monocytogenes EDG-e.
Number of genes downregulated upon ClO2 exposure within the selected role category.
Odd ratios were employed to characterize the representation within each category.
The chi-square test was used to identify the significant association between the number of genes overly inactive and the total number of genes within the selected role category. A P value of <0.05 was used to determine significance.
Genes in two major regulatory networks are differentially expressed under conditions of ClO2 stress.
A significant number of genes under the regulation of the alternative sigma factor σB were differentially expressed. Specifically, a total of 113 genes previously identified as σB dependent (26, 34, 37) were upregulated in cells exposed to ClO2. In total, 51 of the 113 upregulated genes (i) were directly preceded by a σB-dependent promoter or (ii) were in one of 43 operons preceded by a σB-dependent promoter identified as indicating direct σB regulation by Oliver et al. (34). lmo0895, which encodes σB, was upregulated in cells exposed to ClO2 (FC = 1.62; adjusted P value < 0.05). There was an increase in expression of σB regulators, including rsbV (anti-anti-sigma factor encoded by lmo0893) and rsbW (anti-sigma factor encoded by lmo0894); however, they were not significantly differentially expressed as defined by our inclusion criteria. There was an increase in activity for genes regulated by the negative regulator CtsR. The entire putative operon for CtsR (ctsR, mcsA, mcsB, and clpC) was significantly upregulated (adjusted P value < 0.05). A total of 10 genes (mcsA, mcsB, clpC, ynzC, lmo0811, lmo0822, lmo2054, clpB, lmo2205, and clpP) of 42 previously reported as being negatively controlled by CtsR were upregulated (24). Upregulation of 6 of 22 genes identified as indirectly and positively regulated by CtsR (24) was also observed.
TaqMan qRT-PCR confirms selected differentially expressed genes upon exposure of strains to ClO2.
TaqMan qRT-PCR was used to confirm differences in transcript levels of select differentially expressed genes identified by microarray analysis. TaqMan qRT-PCR confirmed statistically significant increases (P value < 0.05) in transcript levels in sigB, lmo0669, lmo1433, clpC, and dnaK in cells exposed to ClO2 on the basis of log copy number (Fig. 2). The increased transcript levels of sigB and clpC support microarray results in that oxidative stress by ClO2 leads to the increased transcriptional activity of both sigB and ctsR and subsequently the σB and CtsR regulons. Transcript levels of lexA were not significantly increased in cells exposed to ClO2 on the basis of qRT-PCR analyses, although a small increase in log copy number was observed (Fig. 2). Limited increases in the lexA log copy number are consistent with fold change values identified by microarray analysis of lexA (FC = 1.61; adjusted P value = 0.044) (see Table S1 in the supplemental material).
FIG 2.
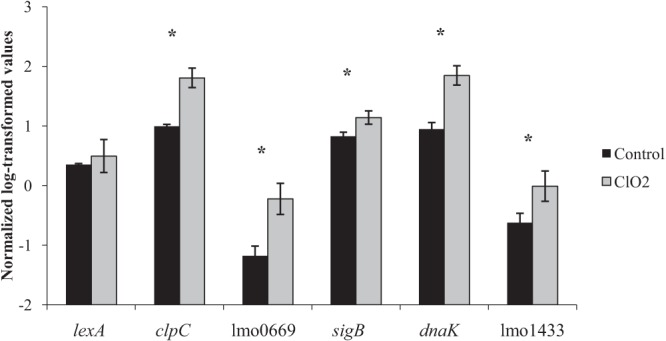
Confirmation of differentially expressed genes by TaqMan qRT-PCR. Transcript levels in lexA, clpC, lmo0669, sigB, dnaK, and lmo1433 were quantified in wild-type strain 10403S by TaqMan qRT-PCR. Transcript levels are expressed as log copy number of cDNA normalized to rpoB. Control samples were not exposed to ClO2; L. monocytogenes was exposed to 300 mg/liter ClO2 for 15 min. Samples were collected and analyzed in triplicate, with standard deviations represented by error bars. Significant differences between control and ClO2-exposed samples were determined by PROC GLM with Dunnett correction; significantly different transcript levels are denoted by an asterisk (P value < 0.05).
σB and CtsR contribute to L. monocytogenes ClO2 survival.
We exposed L. monocytogenes 10403S ΔsigB and ΔctsR strains to 300 mg/ml ClO2 to determine if these regulators affect the survival of L. monocytogenes. The mutant strains were compared to wild-type control cells exposed to 300 mg/liter ClO2 over time (Table 2). There was a significant effect of “strain” (P < 0.001), but there was a not a significant effect of “time” (P = 0.393) or “time*strain” (P = 0.917). At 2.5, 7.5, 10, 15, and 20 min postexposure to 300 mg/liter ClO2, there were significant differences in survival among L. monocytogenes 10403S, ΔsigB, and ΔctsR. Specifically, the ΔsigB and ΔctsR strains were more sensitive to ClO2 than the wild-type cells under the same conditions. This suggests that these regulators play a direct role in ClO2 survival as shown by a significant transcriptional change in the regulons and differences in survival.
DISCUSSION
In this study, we used transcriptomic and phenotypic approaches to investigate the effects of ClO2 on L. monocytogenes. Our data indicate that (i) ClO2 exposure at sublethal levels resulted in differential gene expression of 340 genes, including genes involved in heat shock response, redox reactions, cell replication, and universal stress response, and (ii) stress response regulon networks of σB and CtsR contribute to ClO2 exposure survival. Consistent with other studies investigating L. monocytogenes transcriptional responses to stress (e.g., references 20, 28, and 38), a significant percentage (>11%) of currently identified genes in strain 10403S were differentially expressed. In Bacillus subtilis, oxidative stress induced similar degrees of differential gene expression, with >200 genes upregulated upon exposure to either hydrogen peroxide or paraquat (39). Among the significantly differentially expressed genes, 223 were upregulated and 117 were downregulated. Our data support the hypothesis that L. monocytogenes employs a wide array of stress response systems and that stress response regulators σB and CtsR play direct roles in responding to ClO2 oxidative stress.
Chaperone proteins play a role in oxidative stress management.
Oxidizing agents commonly damage the stability and functionality of proteins. We found 16 genes involved with protein fate activity to be upregulated upon ClO2 exposure. It is likely that increased transcription of peptidase- and protease-based chaperone protein genes (e.g., clpC, clpB, and clpP) is needed for protein degradation and protein recycling and otherwise for repair of proteins damaged by oxidation. Increased activity of genes encoding chaperone proteins (e.g., dnaK, groEL, and groES) indicated a need for maintenance or repair of protein structure damaged by oxidation. These genes are involved in the heat shock response of L. monocytogenes (38), indicating that they have cross-functional stress response utility. Similar activity of heat shock proteins has been observed in E. coli to manage oxidative stress (40). In E. coli, the roles of chaperone repair proteins DnaK, Hsp33, GroEL, and GroES are cross-functional for oxidative and heat shock response activities (41, 42). Increased transcript levels of dnaK, groES, groEL, and clpP have been observed following exposure to H2O2 and chlorine (40). lmo0222, an orthologue to hsp33, was not differentially expressed; however, this specific chaperone has been observed to be controlled posttranslationally (43). A partnership between Hsp33 and DnaK occurs where the oxidized dimers of Hsp33 bind to damaged proteins and, once a redox reaction occurs, transfer the protected substrate protein to DnaK for refolding (42).
Altered activity of cell division genes, a universal stress response, and an SOS response are evident following ClO2 exposure.
An abundance of genes related to cell division and cell macromolecule synthesis were downregulated upon exposure to ClO2 in this study. Downregulation of genes that encode cell division proteins (rodA, ftsW, ftsL, ftsX, and ftsE), septum formation (divIC), and movement (minD) suggests that there is altered activity of the cell division machinery. Genes encoding cell wall structural component assembly, including those encoding teichoic acid, acyl-carrier proteins (lmo0970 to lmo0974), and components for DNA synthesis (genes encoding steps in nucleotide production), also showed decreased transcription. It is tempting to speculate that the absence of significantly differentially expressed genes responsible for cell division indicates stalled cell replication. The decrease in expression levels for genes encoding cell wall manufacturing components bolsters the claim for arrested cell division. While genes involved in cell division and replication were downregulated, universal stress protein (Usp) genes, including lmo0515, lmo1580, and lmo2673, were upregulated. It has been suggested that these three Usp genes are involved with DNA damage repair and the arrest of cell division (37). Their transcription is σB dependent (37), and they aid in acid and oxidative stress survival (44). Usp has been shown to play a key role in survival of E. coli with respect to oxidative species (45). Increased activity of several key SOS response-related genes, including lexA, uvrA, recN, recO, lmo1881, and ynxZ, was also observed here. The role of the SOS regulon and protection of the cell from the reactive species generated through oxidative stress were also observed in B. subtilis (39).
Genes involved with redox reactions help manage ClO2 oxidative stress.
We found increased transcription of thioredoxin, oxidoreductase, superoxide dismutase, catalase, and glutathione reductase genes in cells exposed to ClO2, which outlines the approach of L. monocytogenes to reduce or manage oxidative species. Multiple redox-sensitive transcriptional regulators, including spxA (lmo2191), an ortholog of oxidative stress regulator Spx, were upregulated in this study. In B. subtilis, Spx regulates the thioredoxin-related genes trxA and trxB; thioredoxin aids in oxidative stress management in B. subtilis, working as a substrate for preferential reduction at the expense of NADPH (46). Similarly, we observed increased transcription of thioredoxin genes trxA and lmo2830 as well as lmo1789, which encodes an NADPH oxidoreductase. Further, clpX and clpP, which encode the protease complex ClpXP, which regulates Spx levels (47), were upregulated here, supporting the hypothesis that NADPH-dependent oxidoreductases play roles in managing chlorine dioxide stress. lmo2199 and lmo2200, which have similar levels of homology to the ohrAR operon in B. subtilis, were upregulated as well. OhrA, encoded by ohrA, is involved in organic hydroperoxide resistance in B. subtilis (48). Although OhrR has been identified as a repressor of ohrA, it was observed here that both were upregulated, suggesting that the cell may be regulating an overabundance of OhrA. This same phenomenon was observed in L. monocytogenes under conditions of oxidative stress in the cytosol during intracellular host adaptation (49). Transcription of lmo0613 was significantly increased upon ClO2 exposure. Its orthologue, marA (an oxidoreductase), and its entire operon have been shown to increase resistance to oxidative and antibiotic stress in E. coli (50). In the case of lmo0669, this oxidoreductase has been identified in previous studies as a general stress response protein (26, 35, 37). Fri, a nonheme iron-binding ferritin, encoded by lmo0943, and kat, which encodes catalase, were upregulated as well. Fri has been associated with resistance to hydrogen peroxide, and kat, a catalase gene with a heme group cofactor, has been shown to reduce reactive oxygen species (51, 52). Glutathione has been shown to protect Lactococcus lactis against oxidative stress, specifically, that caused by H2O2 (53). Glutathione reductase reduces glutathione disulfide to glutathione. Increased transcription of an abundance of oxidoreductase genes suggests that the cell may be directing the reactive species toward oxidizing renewable energy targets [e.g., glutathione, NAD(P)H, ATP, etc.] as these are essential for proliferation (53).
σB and CtsR contribute to L. monocytogenes ClO2 survival.
We observed significant transcriptional changes among genes in the σB and CtsR regulatory networks and, to some extent, the HrcA regulatory network upon exposure to ClO2. A total of 113, 16, and 9 genes previously identified as regulated by σB (54), CtsR (55), and HrcA (56), respectively, were upregulated in this study. These three regulons constitute an abundance of stress response genes with overlapping and, in some cases, competing responsibilities for transcriptional control (16). We found that the ΔsigB and ΔctsR mutants had a significantly larger cell population decrease than the wild-type strain. Similar rapid responses were observed in B. subtilis within 10 min of exposure to superoxide and peroxide stressors inducing a notable transcriptional response (39). Stress response genes under direct and indirect control of σB have been found to be differentially expressed during osmotic (35, 57), acid (23, 26, 35), cold (28), and ethanol (26) stress. The CtsR (class three stress gene repressor) regulon negatively regulates heat shock-type proteins; these proteins are important for resistance to high pressure, high temperature, and osmotic stress (58). The HrcA (heat regulation at CIRCE) regulon negatively regulates a separate class of heat shock-type proteins (class I stress response genes). This group includes the genes constituting the dnaK and groESL operons (56), classic, well-characterized chaperone protein systems. A discrepancy occurs between the upregulation of genes directly negatively regulated by CtsR and increased expression of ctsR itself. It is possible that CtsR was being degraded or was otherwise inactive to enlist the stress response genes controlled by this repressor. As inactivation by ClO2 is rapid, as shown by the phenotypic assays described above, and as it is likely that the long-term reactivity of ClO2 is limited by the organic load in the growth medium, the cells may already be trending toward basal expression levels.
Previous studies have demonstrated the ability of ClO2 to inactivate food-borne pathogens on food and food contact surfaces (e.g., reference 8), but this is the first study, to our knowledge, to investigate the effects of ClO2 at the transcriptional level. In this study, we found a substantial number of genes differentially expressed upon sublethal exposure to ClO2, including genes involved in heat shock response and redox reactions. We also found that stress response regulators σB and CtsR directly contribute to ClO2 exposure survival. We observed a clear concentration-dependent effect of ClO2 on survival of stationary-phase L. monocytogenes cells. While oxidation by ClO2 is an immediate reaction, we observed a secondary population decrease, which may result from secondary derivative reactive oxygen species stemming from ClO2 (e.g., ClO2−, Cl−, and ClO3−). We recognize that this study was limited to the transcriptional response of L. monocytogenes to ClO2 in a BHI broth system (as opposed to on produce or food contact surfaces). The volatility of ClO2 results in experimental design challenges. Bacterial cultures were exposed to ClO2 in a BHI broth system, as centrifugation and resuspension of cells in buffer have been observed to alter transcriptional profiles (28). Additionally, increased volatility at higher temperatures was a concern at 37°C; however, lowering the temperature of exposure would have led to differential gene expression from temperature shifts alone. Further analysis should be considered to assess the effects of ClO2 on L. monocytogenes to include (i) multiple times, as only 15 min of exposure to 300 mg/liter was assessed, and (ii) multiple strains from various serotypes and lineages, as only strain 10403S, a 1/2a serotype, was chosen. This initial report provides significant insight into the mechanisms used by L. monocytogenes to respond to and survive oxidative stress from a sanitizer that is increasingly being used in the agricultural commodities and food industries. A thorough understanding of bactericidal mechanisms is pivotal in understanding the utility of sanitizers. This understanding will allow the synergistic application of multiple sanitizer agents with different modes of actions to boost the efficacy and optimization of future sanitizers, both of which will lead to more effective approaches to inactivation of L. monocytogenes in food processing environments.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Purdue University College of Agriculture Research Funds and USDA-ARS project number 1935-42000-049-00D.
We thank Kathryn J. Boor and Martin Wiedmann at Cornell University for L. monocytogenes strains 10403S, ΔsigB, and ΔctsR and for the microarray slides. We also thank the laboratory of Jennifer Freeman for the use of the Genepix 4000B scanner and technical support.
Footnotes
Published ahead of print 7 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00004-14.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.p11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver HF, Wiedmann M, Boor KJ. 2007. Environmental reservoir and transmission into the mammalian host, p 111–137 In Goldfine H, Shen H. (ed), Listeria monocytogenes: pathogenesis and host response. Springer Science, New York, NY [Google Scholar]
- 3.Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, Wiedmann M. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458–4467. 10.1128/AEM.70.8.4458-4467.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thimothe J, Nightingale KK, Gall K, Scott VN, Wiedmann M. 2004. Tracking of Listeria monocytogenes in smoked fish processing plants. J. Food Prot. 67:328–341 [DOI] [PubMed] [Google Scholar]
- 5.Hoelzer K, Sauders BD, Sanchez MD, Olsen PT, Pickett MM, Mangione KJ, Rice DH, Corby J, Stich S, Fortes ED, Roof SE, Grohn YT, Wiedmann M, Oliver HF. 2011. Prevalence, distribution, and diversity of Listeria monocytogenes in retail environments, focusing on small establishments and establishments with a history of failed inspections. J. Food Prot. 74:1083–1095. 10.4315/0362-028X.JFP-10-567 [DOI] [PubMed] [Google Scholar]
- 6.Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P, Frankel G. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12:2385–2397. 10.1111/j.1462-2920.2010.02297.x [DOI] [PubMed] [Google Scholar]
- 7.López-Gálvez F, Allende A, Truchado P, Martínez-Sánchez A, Tudela JA, Selma MV, Gil MI. 2010. Suitability of aqueous chlorine dioxide versus sodium hypochlorite as an effective sanitizer for preserving quality of fresh-cut lettuce while avoiding by-product formation. Postharvest. Biol. Tec. 55:53–60. 10.1016/j.postharvbio.2009.08.001 [DOI] [Google Scholar]
- 8.Han Y, Linton RH, Nielsen SS, Nelson PE. 2001. Reduction of Listeria monocytogenes on green peppers (Capsicum annuum L.) by gaseous and aqueous chlorine dioxide and water washing and its growth at 7 degrees C. J. Food Prot. 64:1730–1738 [DOI] [PubMed] [Google Scholar]
- 9.Ryu JH, Beuchat LR. 2005. Biofilm formation and sporulation by Bacillus cereus on a stainless steel surface and subsequent resistance of vegetative cells and spores to chlorine, chlorine dioxide, and a peroxyacetic acid-based sanitizer. J. Food Prot. 68:2614–2622 [DOI] [PubMed] [Google Scholar]
- 10.Mahmoud BS, Vaidya NA, Corvalan CM, Linton RH. 2008. Inactivation kinetics of inoculated Escherichia coli O157:H7, Listeria monocytogenes and Salmonella Poona on whole cantaloupe by chlorine dioxide gas. Food Microbiol. 25:857–865. 10.1016/j.fm.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 11.Vaid R, Linton RH, Morgan MT. 2010. Comparison of inactivation of Listeria monocytogenes within a biofilm matrix using chlorine dioxide gas, aqueous chlorine dioxide and sodium hypochlorite treatments. Food Microbiol. 27:979–984. 10.1016/j.fm.2010.05.024 [DOI] [PubMed] [Google Scholar]
- 12.Ogata N. 2007. Denaturation of protein by chlorine dioxide: oxidative modification of tryptophan and tyrosine residues. Biochemistry 46:4898–4911. 10.1021/bi061827u [DOI] [PubMed] [Google Scholar]
- 13.Bakhmutova-Albert EV, Margerum DW, Auer JG, Applegate BM. 2008. Chlorine dioxide oxidation of dihydronicotinamide adenine dinucleotide (NADH). Inorg. Chem. 47:2205–2211. 10.1021/ic7019022 [DOI] [PubMed] [Google Scholar]
- 14.Smigic N, Rajkovic A, Arneborg N, Siegumfeldt H, Devlieghere F, Nielsen DS. 2012. Analysis of intracellular pH in Escherichia coli O157:H7 to determine the effect of chlorine dioxide decontamination. Food Anal. Methods 5:327–331. 10.1007/s12161-011-9295-0 [DOI] [Google Scholar]
- 15.Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527–543. 10.1128/MMBR.69.4.527-543.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaturongakul S, Raengpradub S, Palmer ME, Bergholz TM, Orsi RH, Hu Y, Ollinger J, Wiedmann M, Boor KJ. 2011. Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors σB, σC, σH, and σL in Listeria monocytogenes. Appl. Environ. Microbiol. 77:187–200. 10.1128/AEM.00952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver HF, Orsi RH, Wiedmann M, Boor KJ. 2010. Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76:4216–4232. 10.1128/AEM.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giotis ES, Julotok M, Wilkinson BJ, Blair IS, McDowell DA. 2008. Role of sigma B factor in the alkaline tolerance response of Listeria monocytogenes 10403S and cross-protection against subsequent ethanol and osmotic stress. J. Food Prot. 71:1481–1485 [DOI] [PubMed] [Google Scholar]
- 19.Skandamis PN, Yoon Y, Stopforth JD, Kendall PA, Sofos JN. 2008. Heat and acid tolerance of Listeria monocytogenes after exposure to single and multiple sublethal stresses. Food Microbiol. 25:294–303. 10.1016/j.fm.2007.10.008 [DOI] [PubMed] [Google Scholar]
- 20.Bergholz TM, Bowen B, Wiedmann M, Boor KJ. 2012. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 78:2602–2612. 10.1128/AEM.07658-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garner MR, James KE, Callahan MC, Wiedmann M, Boor KJ. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl. Environ. Microbiol. 72:5384–5395. 10.1128/AEM.00764-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005–2009 [PubMed] [Google Scholar]
- 23.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Raengpradub S, Schwab U, Loss C, Orsi RH, Wiedmann M, Boor KJ. 2007. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and sigma B in Listeria monocytogenes. Appl. Environ. Microbiol. 73:7967–7980. 10.1128/AEM.01085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trinetta V, Vaidya N, Linton R, Morgan M. 2011. Evaluation of chlorine dioxide gas residues on selected food produce. J. Food Sci. 76:T11–T15. 10.1111/j.1750-3841.2010.01911.x [DOI] [PubMed] [Google Scholar]
- 26.Raengpradub S, Wiedmann M, Boor KJ. 2008. Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74:158–171. 10.1128/AEM.00951-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. Craig Venter Institute. 2006. The Institute for Genomic Research standard operating procedure M007: microbial RNA aminoallyl labeling for microarrays. http://pfgrc.jcvi.org/index.php/microarray/protocols.html
- 28.Chan YC, Boor KJ, Wiemdann M. 2007. σB-Dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl. Environ. Microbiol. 73:6019–6029. 10.1128/AEM.00714-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Craig Venter Institute. 2006. The Institute for Genomic Research Standard operating procedure M008: hybridization of labeled DNA and CDNA probes. http://pfgrc.jcvi.org/index.php/microarray/protocols.html
- 30.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3 [DOI] [PubMed] [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700–2707. 10.1093/bioinformatics/btm412 [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273. 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- 34.Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour SW, Filiatrault MJ, Wiedmann M, Boor KJ. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. 10.1186/1471-2164-10-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sue D, Fink D, Wiedmann M, Boor KJ. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843–3855. 10.1099/mic.0.27257-0 [DOI] [PubMed] [Google Scholar]
- 36.Hahne H, Mader U, Otto A, Bonn F, Steil L, Bremer E, Hecker M, Becher D. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882. 10.1128/JB.01106-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hain T, Hossain H, Chatterjee SS, Machata S, Volk U, Wagner S, Brors B, Haas S, Kuenne CT, Billion A, Otten S, Pane-Farre J, Engelmann S, Chakraborty T. 2008. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σB regulon. BMC Microbiol. 8:20. 10.1186/1471-2180-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Veen S, Hain T, Wouters JA, Hossain H, de Vos WM, Abee T, Chakraborty T, Wells-Bennik MH. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153:3593–3607. 10.1099/mic.0.2007/006361-0 [DOI] [PubMed] [Google Scholar]
- 39.Mostertz J, Scharf C, Hecker M, Homuth G. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497–512. 10.1099/mic.0.26665-0 [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, Tortorello ML, Zhang W. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75:6110–6123. 10.1128/AEM.00914-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. 2008. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135:691–701. 10.1016/j.cell.2008.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter J, Linke K, Jatzek A, Jakob U. 2005. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell 17:381–392. 10.1016/j.molcel.2004.12.027 [DOI] [PubMed] [Google Scholar]
- 43.Graf PC, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. 2004. Activation of the redox-regulated chaperone Hsp33 by domain unfolding. J. Biol. Chem. 279:20529–20538. 10.1074/jbc.M401764200 [DOI] [PubMed] [Google Scholar]
- 44.Wemekamp-Kamphuis HH, Wouters JA, de Leeuw PP, Hain T, Chakraborty T, Abee T. 2004. Identification of sigma factor sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457–3466. 10.1128/AEM.70.6.3457-3466.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nachin L, Nannmark U, Nystrom T. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187:6265–6272. 10.1128/JB.187.18.6265-6272.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scharf C, Riethdorf S, Ernst H, Engelmann S, Volker U, Hecker M. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano S, Kuster-Schock E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608. 10.1073/pnas.2235180100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134–4141. 10.1128/JB.183.14.4134-4141.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323–1338. 10.1128/IAI.74.2.1323-1338.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abram F, Starr E, Karatzas KA, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O'Byrne CP. 2008. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74:6848–6858. 10.1128/AEM.00442-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen KN, Larsen MH, Gahan CG, Kallipolitis B, Wolf XA, Rea R, Hill C, Ingmer H. 2005. The Dps-like protein Fri of L. monocytogenes promotes stress tolerance and intrucellular multiplication in macrophase-like cells. Microbiology 151:925–933. 10.1099/mic.0.27552-0 [DOI] [PubMed] [Google Scholar]
- 52.Rea R, Hill C, Gahan CG. 2005. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71:8314–8322. 10.1128/AEM.71.12.8314-8322.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Hugenholtz J, Abee T, Molenaar D. 2003. Glutathione protects Lactococcus lactis against oxidative stress. Appl. Environ. Microbiol. 69:5739–5745. 10.1128/AEM.69.10.5739-5745.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. 2003. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185:5722–5734. 10.1128/JB.185.19.5722-5734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nair S, Derre I, Msadek T, Gaillot O, Berche P. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800–811. 10.1046/j.1365-2958.2000.01752.x [DOI] [PubMed] [Google Scholar]
- 56.Hu Y, Oliver HF, Raengpradub S, Palmer ME, Orsi RH, Wiedmann M, Boor KJ. 2007. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and σB in Listeria monocytogenes. Appl. Environ. Microbiol. 73:7981–7991. 10.1128/AEM.01281-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker LA, Cetin MS, Hutkins RW, Benson AK. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karatzas KA, Wouters JA, Gahan CG, Hill C, Abee T, Bennik MH. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227–1238. 10.1046/j.1365-2958.2003.03636.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.