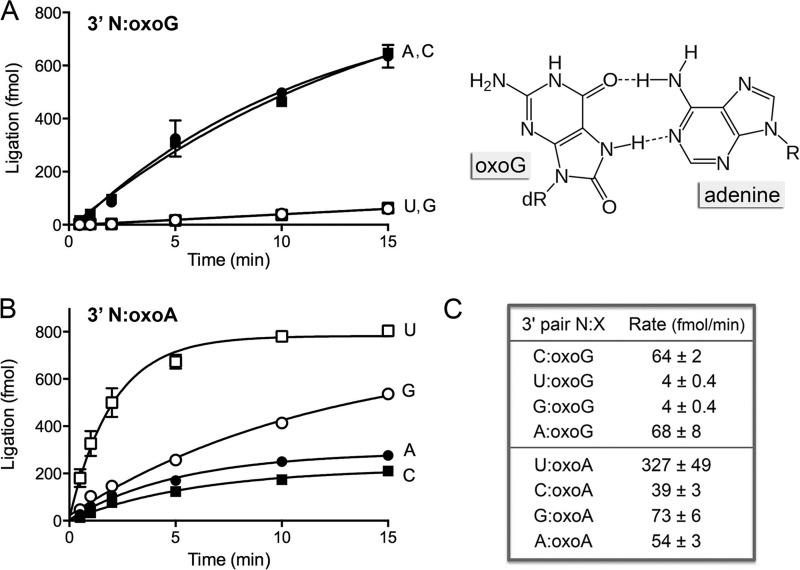
FIG 3.
Effect of 8-oxopurines in the template strand at the nick 3′-OH end. (A) The substrates for sealing were versions of the nicked duplex shown in Fig. 1A, in which the X nucleobase of the 3′-OH N-X end was either oxoG (A) or oxoA (B). The chemical structure of the A-oxoG pair with oxoG in the syn conformation is shown at the right in panel A; hydrogen-bonding contacts are indicated by dashed lines. dR, deoxyribose. Ligase reactions were performed as described in Materials and Methods. The yields of the sealed 36-mer RNApDNA product are plotted as a function of time; the experiments are presented in sets of four, with the N nucleobase (specified to the right of each curve) being the variable within the set. Each datum is the average of at least 3 separate experiments ± SEM. (C) The initial rates of sealing of the indicated 3′ N-oxopurine nicks were calculated by linear regression in Prism.