FIG 1.
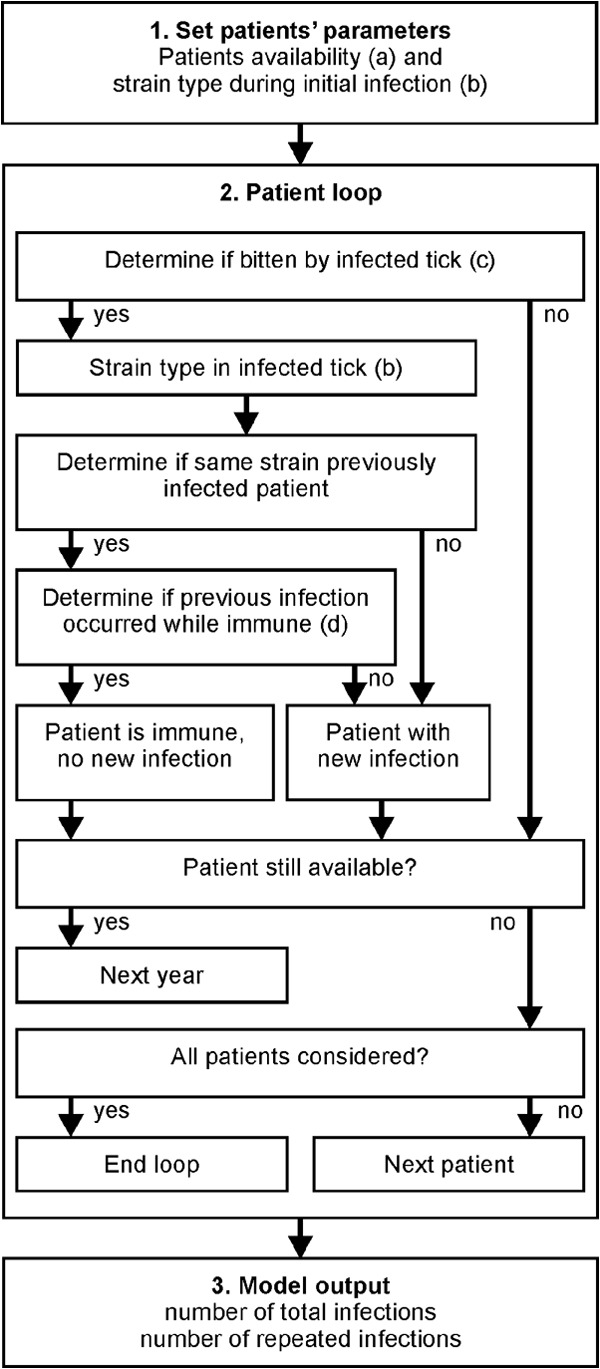
Schematic representation of the stochastic simulation model. (1) Set patients' parameters. The model uses the patient availability from the appropriate data set (limited or extended availability) (a) and determines the strain in the initial infection of each patient from the appropriated strain probability distribution (data set 1, 2, or 3) (b). (2) Patient loop. For each patient, the model first determines, for each year that the patient is available, if the patient is bitten by an infected tick (c). If the patient is exposed to an infected tick, the model determines the B. burgdorferi strain carried by the infecting tick (b). If the patient has previously been infected by the strain in the infecting tick within the duration of strain-specific immunity (d), that tick bite does not result in a new infection. If the patient is not immune, a new infection is recorded. The process is repeated until the patient is no longer available, at which point the model simulates the next patient until the infections in all patients have been simulated. (3) Model output. The model saves the total number of infections and the number of patients that have been infected by the same strain multiple times for each iteration of the model. For each combination of parameters (Table 1), data from 10,000 iterations are recorded.
