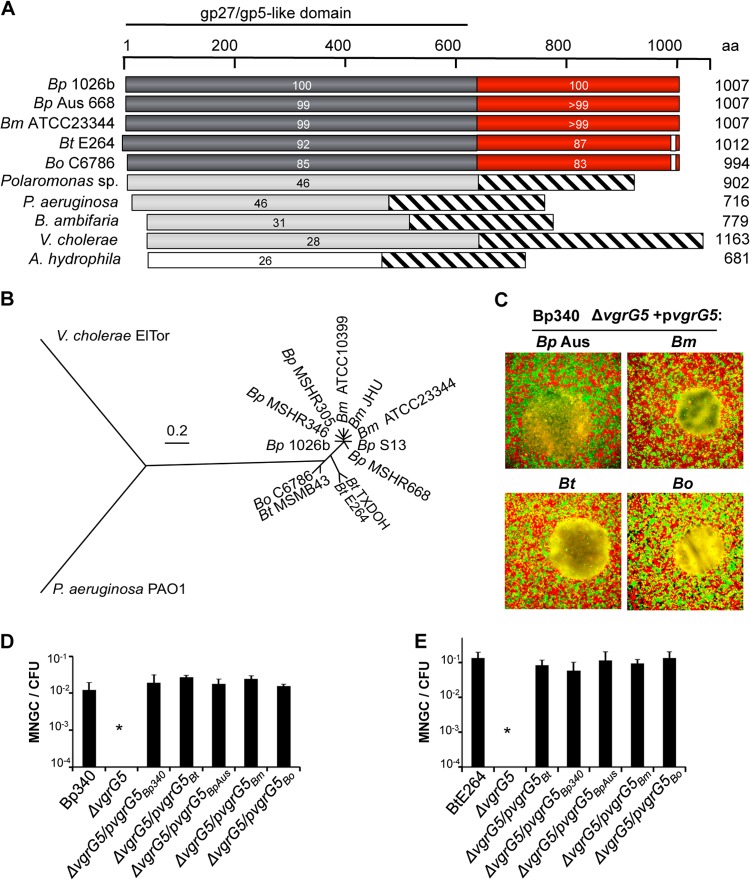
FIG 2.
vgrG5 alleles from Pseudomallei group Burkholderia species are functionally interchangeable. (A) Alignment of VgrG proteins by BLAST. Gray bars, amino-terminal VgrG “core” region with homology to the gp27/gp5 T4 bacteriophage tail spike complex proteins; red bars, conserved CTD; striped bars, protein regions with no similarities. The boundary of the CTD was assigned based on the alignment of the Burkholderia VgrG5 sequence to the sequences of VgrG proteins in other species. The percentage of identity with the B. pseudomallei 1026b VgrG5 coding sequence is given for each sequence diagramed here. Bp, B. pseudomallei; Bm, B. mallei; Bt, B. thailandensis; Bo, B. oklahomensis. (B) Phylogenetic tree based on comparisons of sequences of VgrG5 proteins from an expanded set of Burkholderia isolates and of VgrG and VgrG1 proteins from V. cholerae and P. aeruginosa, respectively. Analysis was based on the JTT matrix-based model (62) and was conducted in MEGA5 (63). (C) MNGC assays. HEK293 cells were infected with a Bp340 ΔvgrG5 mutant complemented with a plasmid expressing vgrG5 from the Australian B. pseudomallei strain MSHR668 (Bp Aus), B. mallei ATCC 23344, B. thailandensis E264, or B. oklahomensis C6786. (D and E) Numbers of MNGCs per CFU 18 h after infection of HEK293 cells with B. pseudomallei Bp340 (D) or B. thailandensis E264 (E) containing ΔvgrG5 deletions and complementing plasmids as indicated. No fusion activity was observed in ΔvgrG5 strains containing the empty vector. Values are means ± SD for 3 independent experiments (*, P < 0.05).