Abstract
Since the first step of the infection process is colonization of the host, it is important to understand how Escherichia coli pathogens successfully colonize the intestine. We previously showed that enterohemorrhagic O157:H7 strain E. coli EDL933 colonizes a niche in the streptomycin-treated mouse intestine that is distinct from that of human commensal strains, which explains how E. coli EDL933 overcomes colonization resistance imparted by some, but not all, commensal E. coli strains. Here we sought to determine if other E. coli pathogens use a similar strategy. We found that uropathogenic E. coli CFT073 and enteropathogenic E. coli E2348/69 occupy intestinal niches that are distinct from that of E. coli EDL933. In contrast, two enterohemorrhagic strains, E. coli EDL933 and E. coli Sakai, occupy the same niche, suggesting that strategies to prevent colonization by a given pathotype should be effective against other strains of the same pathotype. However, we found that a combination of commensal E. coli strains that can prevent colonization by E. coli EDL933 did not prevent colonization by E. coli CFT073 or E. coli E2348/69. Our results indicate that development of probiotics to target multiple E. coli pathotypes will be problematic, as the factors that govern niche occupation and hence stable colonization vary significantly among strains.
INTRODUCTION
Pathogenic strains of Escherichia coli, without exception, must first colonize the host gastrointestinal tract before causing disease (1). Freter (2) postulated that successful competition for nutrients allows intestinal bacteria to colonize, which we define as the ability to achieve and maintain a stable population without reintroduction. Freter's nutrient niche hypothesis theoretically explains the succession of community members of the intestinal microbiota, as well as the ability of enteric pathogens to overcome colonization resistance and thereby invade the microbiota (3). How invading pathogens compete for nutrients with the established microbial residents is an open question.
Recently, we developed a colonization resistance model in streptomycin-treated mice, in which a given strain cannot colonize mice that are precolonized with the same E. coli strain (4, 5). We demonstrated that several human-derived E. coli strains are able to overcome colonization resistance imparted by other E. coli strains and thereby cocolonize the intestine. The results establish that various commensals, as well as enterohemorrhagic E. coli (EHEC) strain O157:H7, occupy distinct niches in the intestine and therefore can properly be described as ecotypes and pathotypes, respectively.
A series of previous studies from our laboratories indicate that in the mouse intestine different pathogenic and commensal E. coli strains each use a different set of approximately 6 sugars of the more than 18 sugars that E. coli potentially is capable of using in vitro (6–11). This led us to hypothesize that invading E. coli pathogens overcome colonization resistance by taking advantage of nutrients that are available because they are not used by resident commensal E. coli strains (8). While this is an attractive model and seemingly is correct, we subsequently found that the intestine selects for flhD mutants of E. coli K-12, which are superior colonizers not by virtue of using different sugars but rather by using the same sugars better than its wild-type parent (12, 13). Intriguingly, the mouse intestine also selects for envZ missense mutants that hyperphosphorylate OmpR, causing them to have an altered outer membrane protein profile, to grow more slowly in vitro on cecal mucus as well as several mono- and disaccharides, and to outcompete their wild-type parent during colonization of the streptomycin-treated mouse intestine (14). Furthermore, we found that the envZ mutants occupy an intestinal niche in which galactose is available but from which the wild-type parent is excluded (14). To explain these results, we invoked the “restaurant” hypothesis (14), which states that sugars are made available locally to commensal E. coli by polysaccharide-degrading anaerobes within the mixed biofilms that E. coli inhabits (15). Collectively, these competitive strategies potentially allow different ecotypes and pathotypes to occupy different niches.
Competition between invading pathogens and the microbiota undergirds the concept of probiotics as a preventative measure. That there is stiff competition between strains for nutrients in the intestine emphasizes the potential of probiotics to limit the availability of nutrients required by pathogens to colonize (9). Indeed, we recently showed that while no single commensal E. coli strain can exert colonization resistance against E. coli EDL933 (the prototypical O157:H7 strain), a mixture of several commensals could (4). By considering the profiles of sugars utilized by several commensal E. coli, we found two strains that overlap most but not all of the sugar catabolism capacity of E. coli EDL933 and together are able to prevent the pathogen from colonizing (5). One of these two strains, E. coli Nissle 1917, has long been recognized as having probiotic potential against dysentery and traveler's diarrhea (16), and the other, E. coli HS, is well established as an excellent colonizer of primates (17). It is interesting to consider the possibility that appropriate mixtures of commensal E. coli strains might consume the nutrients needed by enteric pathogens, in general, to colonize and thereby prevent disease. While preventing colonization by E. coli EDL933 is a promising step in the development of probiotics, this approach has not been tested for other E. coli pathotypes, nor is it known whether different pathotypes occupy distinct niches in the intestine.
Here we tested whether uropathogenic E. coli CFT073 (UPEC), enteropathogenic E. coli E2348/69 (EPEC), and the enterohemorrhagic E. coli EDL933 (EHEC) occupy the same or different niches in the streptomycin-treated mouse intestine. Furthermore, we determined whether the combination of commensal strains previously shown to exert colonization resistance to E. coli EDL933 (5) could also prevent colonization by the pathogens E. coli CFT073 and E. coli E2348/69. The results demonstrate that different E. coli pathotypes indeed occupy distinct intestinal niches and commensals that prevent colonization by one pathotype do not prevent colonization by others. We discuss the implication of these results for designing strategies to prevent colonization by enteric pathogens.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. All strains were made Strr (streptomycin resistant) to allow them to colonize streptomycin-treated mice. In addition, the Strr strains were also made either Nalr (nalidixic acid resistant) or Rifr (rifampin resistant) to facilitate plate counting of specific populations in streptomycin-treated mouse colonization experiments, as described previously (5, 6, 10). Each of the isogenic Strr, Strr Nalr, and Strr Rifr strains were competed against one another in control experiments and were found to cocolonize mice, which confirms that the selectable markers did not impact their relative fitness for colonization (references 5, 6, and 10 and data not shown). E. coli EDL933 is the prototype strain of EHEC strain O157:H7 (18). E. coli Sakai was isolated from contaminated vegetables during a massive outbreak in Japan and is serotypically identical to E. coli EDL933 (19). E. coli CFT073 is the sequenced prototype UPEC strain and readily colonizes the gastrointestinal (GI) tract (20), and E. coli E2348/69 is the sequenced prototype EPEC strain (21). E. coli Nissle 1917 (16) and E. coli HS (17) are commensal human isolates. To facilitate plate counting of E. coli HS when colonized with more than one other strain, we used E. coli HS Strr uxaC::kanR because it provides an alternative selectable marker (kanamycin resistance), but loss of uronic isomerase and hence the ability to grow on hexuronic acids does not affect its colonizing ability, as it cocolonizes with its wild-type parent E. coli HS Strr Nalr (5).
TABLE 1.
E. coli strains used in this study
| E. coli strain | Relevant genotype or description | Source or reference |
|---|---|---|
| CFT073 | Wild-type O6:H1 | 20 |
| CFT073 Strr | Spontaneous Strr | Karen Krogfelt |
| CFT Strr Nalr | Spontaneous Strr Nalr | This study |
| EDL933 | Wild-type O157:H7 | Allison O'Brien |
| EDL933 Strr | Spontaneous Strr | 10 |
| EDL 933 Strr Rifr | Spontaneous Strr Rifr | 10 |
| E2348/69 | Wild-type O127:H6 | Jim Nataro |
| E2348/69 Strr | Spontaneous Strr | This study |
| E2348/69 Strr Nalr | Spontaneous Strr Nalr | This study |
| HS | Wild type | Dave Rasko |
| HS Strr | Spontaneous Strr | 5 |
| HS Strr ΔuxaC::kan | Strr ΔuxaC::kan | 5 |
| Nissle 1917 | Wild type | Dean Hamer |
| Nissle 1917 Strr | Spontaneous Strr | 6 |
| Nissle 1917 Strr Rifr | Spontaneous Strr Rifr | 6 |
| Sakai | Wild-type O157:H7 | Beth Whitman |
| Sakai Strr | Spontaneous Strr | This study |
| Sakai Strr Nalr | Spontaneous Strr Nalr | This study |
Streptomycin-treated mouse model.
The streptomycin-treated mouse model is used extensively to study the colonization of Escherichia coli in the mouse large intestine (22). Briefly, three 6-week-old CD-1 mice were obtained from Charles River Laboratories and given streptomycin sulfate (5 g/liter) in their drinking water for 24 h. Streptomycin sulfate selectively eliminates the resident facultative anaerobes in the intestine with little disruption of the anaerobe community, opening a niche for introduced E. coli strains to colonize (23). Following treatment with streptomycin sulfate, the mice were starved of food and water for 14 h and then fed approximately 105 CFU of E. coli strain(s) in 1 ml of 20% sucrose (day 0). Following consumption of the bacterial suspension, food and water (5 g/liter streptomycin sulfate) were given to the mice ad libitum for the duration of the experiment. After 10 days, food and water again were withheld for 14 h, and the mice were fed approximately 105 CFU of a challenge strain. Fecal samples were taken after 5 h and 24 h and then every other day thereafter for 21 days. Antibiotic resistance was used to distinguish the strains in fecal plate counts. Feces were homogenized in 1% tryptone, serially diluted, and plated on lactose-MacConkey agar supplemented with the appropriate antibiotics, as follows: streptomycin sulfate (100 mg/ml), nalidixic acid (50 mg/ml), rifampin (50 mg/ml), and kanamycin (40 mg/ml). Each colonization experiment was repeated, and the values for the six mice (or more) were averaged. The log10 CFU/g feces was determined for each strain at each time point (± the standard error), and a Student t test was performed. A difference of ≥101 CFU/g feces between strains in all cases was statistically significant (P < 0.05).
Enumeration of E. coli strains from mouse intestinal mucus.
Both E. coli HS ΔuxaC::kan Strr Kanr and E. coli Nissle 1917 Strr Rifr were fed to sets of three streptomycin-treated mice and allowed to colonize for 10 days. On day 10, 105 CFU of E. coli E2348/69 Strr Nalr (EPEC) or E. coli CFT073 Strr Nalr (UPEC) was fed to individual sets of three precolonized mice and cocolonization was monitored. After 5 days, we confirmed that all three organisms had cocolonized in the mice, and then the mice were sacrificed, the intestinal tract was removed, sectioned, opened, and gently washed, the mucosal layer of each section was collected, and the population of each strain of E. coli was quantified as previously described (4, 24). Prior to removing the intestinal tract, the bladder of each mouse was removed and placed in 5 ml HEPES-Hanks buffer (pH 7.4) and analyzed for the presence of any of the experimentally introduced E. coli strains. Briefly, the bladder and its contents were homogenized by vortexing, followed by serial dilution in 1% tryptone broth and plating onto MacConkey agar containing the appropriate antibiotics for each strain.
RESULTS
Commensal E. coli cannot prevent colonization by UPEC and EPEC.
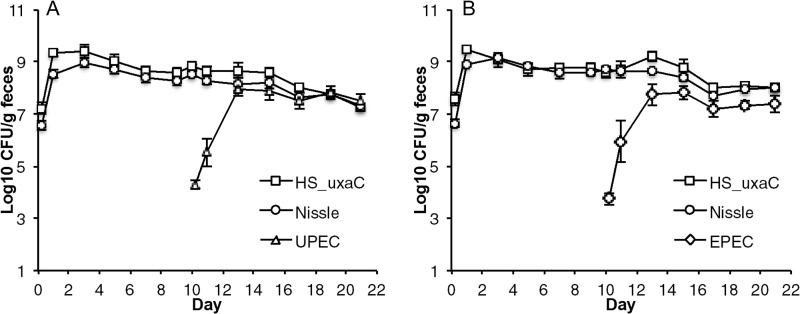
If it is true that invading pathogens must overcome colonization resistance by competing with the microbiota for intestinal niches, then commensal strains that occupy all preferred niches would prevent colonization by pathogens. Previously, we showed that mixtures of human commensal E. coli ecotypes (e.g., E. coli HS and E. coli Nissle 1917) could prevent EHEC strain EDL933 from colonizing the streptomycin-treated mouse intestine (4, 5). If it is true that other E. coli pathogens prefer the same intestinal niches as EHEC, then E. coli HS and E. coli Nissle 1917 should exert colonization resistance against those pathogens. To test this possibility, we precolonized streptomycin-treated mice by feeding them 105 CFU each of E. coli HS and E. coli Nissle 1917, which both colonized at between 108 and 109 CFU per g of feces (Fig. 1). On day 10 of the experiments, mice that were precolonized with E. coli HS and E. coli Nissle 1917 were challenged by association (i.e., feeding) with a third strain, either 105 CFU of E. coli CFT073, a UPEC strain, or 105 CFU of E. coli E2348/69, an EPEC strain. Both UPEC (Fig. 1A) and EPEC (Fig. 1B) grew from low to high numbers in competition with precolonized E. coli HS and E. coli Nissle 1917, reaching populations between 107 and 108 CFU per g of feces by day 13 of the experiments. These results indicate that UPEC and EPEC occupy at least one intestinal niche in mice that is not occupied by either E. coli HS or E. coli Nissle 1917. The results do not determine whether or not the additional niches occupied by UPEC and EPEC are the same, but it is clear that no single strategy for stimulating colonization resistance against more than one E. coli pathotype will be effective.
FIG 1.
UPEC and EPEC colonization of mice precolonized with a combination of commensal E. coli strains HS and Nissle 1917. (A) Two sets of 3 mice were fed 105 CFU of E. coli HS Strr ΔuxaC::kan and E. coli Nissle 1917 Strr Rifr and 10 days later were fed 105 CFU of E. coli CFT073 Strr Nalr (UPEC). (B) Two sets of 3 mice were fed 105 CFU of E. coli HS Strr ΔuxaC::kan and E. coli Nissle 1917 Strr Rifr and 10 days later were fed 105 CFU of E. coli E2348/69 Strr Nalr (EPEC).
Localization of E. coli HS, E. coli Nissle 1917, E. coli EPEC, and E. coli UPEC in the streptomycin-treated mouse GI tract.
The ability of EPEC strain E2348/69 and UPEC strain CFT073 to cocolonize mice that were precolonized by the human commensal strains E. coli HS and E. coli Nissle 1917 (Fig. 1) suggests that the pathogenic strains are competing for different niches. While E. coli HS and E. coli Nissle 1917 have been previously shown to colonize the streptomycin-treated mouse large intestine, primarily the cecum (20), the two pathogenic strains of E. coli, EPEC and UPEC, have not been correlated to any specific region of the streptomycin-treated mouse GI tract. It may be that the pathogenic strains prefer to occupy the small intestine while E. coli HS and Nissle1917 colonize the large intestine. To answer this question, the entire mouse GI tract was excised and cut into sections corresponding to the jejunum, ileum, cecum, and colon, each section was cut open and gently washed, and the mucus layer was collected by scraping; then, the population of each E. coli strain was enumerated by plate counting of the mucus samples (Table 2). When E. coli HS, E. coli Nissle 1917, and EPEC were competed against each other, roughly the same number of each bacterium was recovered from each section of the GI tract (Table 2). The same results were observed when E. coli HS, E. coli Nissle 1917, and UPEC were competed against each other, with E. coli HS showing a 2-fold increase within the jejunum. The numbers of E. coli recovered from each section of the intestinal system were highest within the cecal and colonic mucosal layers. No inflammation was noted in any of the animals colonized with UPEC or EPEC. In addition, none of the strains could be recovered from the bladder (data not shown). These results are in agreement with studies of other pathogens that indicate that the streptomycin-treated mouse is a colonization model, not a pathogenesis model (4, 8). It was noted previously that E. coli EDL933 colonizes the streptomycin-treated mouse cecum in the highest numbers, and histology showed that all tissues were normal (25). Our results show that EPEC strain E2348/69 colonizes the colon in the highest numbers and UPEC strain CFT073 colonizes the GI tract in the highest numbers in the cecum but does not infect the urinary tract.
TABLE 2.
Population distribution of E. coli strains HS, Nissle, and E2348/69 (EPEC) or CFT073 (UPEC) within the small intestinal (jejunum), ileal, cecal, and colonic mucus layers
| Expt and location | Mean log10 CFU ± SEMa for E. coli strain: |
|||
|---|---|---|---|---|
| HS | Nissle | E2348/69 | CFT073 | |
| E2348/69 cocolonization | ||||
| Jejunumb | 4.7 ± 0.1 | 4.2 ± 0.7 | 4.2 ± 0.9 | |
| Ileum | 4.2 ± 0.1 | 3.8 ± 0.1 | 3.8 ± 0.2 | |
| Cecum | 6.1 ± 0.2 | 6.5 ± 0.2 | 5.8 ± 0.3 | |
| Colon | 6.0 ± 0.5 | 6.8 ± 0.1 | 6.4 ± 0.6 | |
| Fecesc | 6.9 ± 0.9 | 7.3 ± 0.7 | 6.3 ± 0.2 | |
| CFT073 cocolonization | ||||
| Jejunumb | 4.7 ± 0.3 | 2.8 ± 0.6 | 2.4 ± 0.3 | |
| Ileum | 3.9 ± 0.4 | 3.9 ± 0.7 | 3.5 ± 1.1 | |
| Cecum | 5.1 ± 0.2 | 5.4 ± 0.4 | 5.5 ± 0.6 | |
| Colon | 5.7 ± 0.2 | 4.8 ± 0.5 | 4.7 ± 0.7 | |
| Fecesc | 6.4 ± 0.1 | 6.1 ± 0.3 | 6.2 ± 0.5 | |
Data from sets of 3 mice. SEM, standard error of the mean.
Mucus taken from below the stomach to the proximal ileum.
Fecal counts are in CFU/gram of feces collected on the day of intestinal sampling, as described in the text.
Two EHEC strains occupy the same intestinal niche.
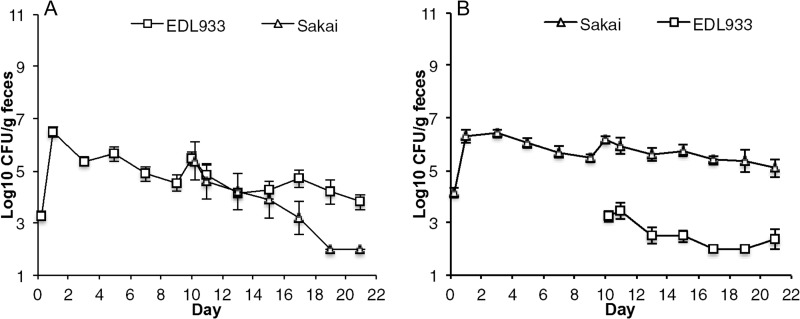
Different E. coli pathotypes share a “backbone” genome but have distinctly different gene complements (26–28). These genetic differences could allow various pathogens to occupy distinct niches in the intestine by bestowing different physiological traits that promote different strain-strain interactions in the intestine. Alternatively, genetically similar strains might be expected to occupy the same niche in the intestine because they express the same physiological traits. Previously, we showed that several E. coli ecotypes exert colonization resistance against isogenic strains (4). So, we sought to determine if the highly similar EHEC strains E. coli EDL933 and E. coli Sakai occupy the same or different niches (Fig. 2). These EHEC strains share >99.9% sequence identity in their protein-coding genes (29), and the relatively minor genomic differences between them can be attributed to how the two genomes were annotated (28). Mice were precolonized with105 CFU of E. coli EDL933 by feeding and on day 10 challenged by association with 105 CFU E. coli Sakai (Fig. 2A). E. coli Sakai failed to grow to higher numbers and over the next 10 days of the experiment was almost eliminated from the intestine. In a reciprocal experiment, mice were precolonized with E. coli Sakai and on day 10 challenged by association with 105 CFU of E. coli EDL933 (Fig. 2B). Again, E. coli EDL933 failed to grow to higher numbers and was almost eliminated from the intestine. These results indicate that the closely related EHEC strains exert colonization resistance against one another and occupy the same intestinal niches.
FIG 2.
Competitive colonization between EHEC strains. (A) Two sets of 3 mice were fed 105 CFU of E. coli EDL933 Strr Rifr and 10 days later were fed 105 CFU of E. coli Sakai Strr Nalr. (B) The reciprocal experiment was conducted.
UPEC and EPEC occupy intestinal niches that are distinct from that of EHEC.
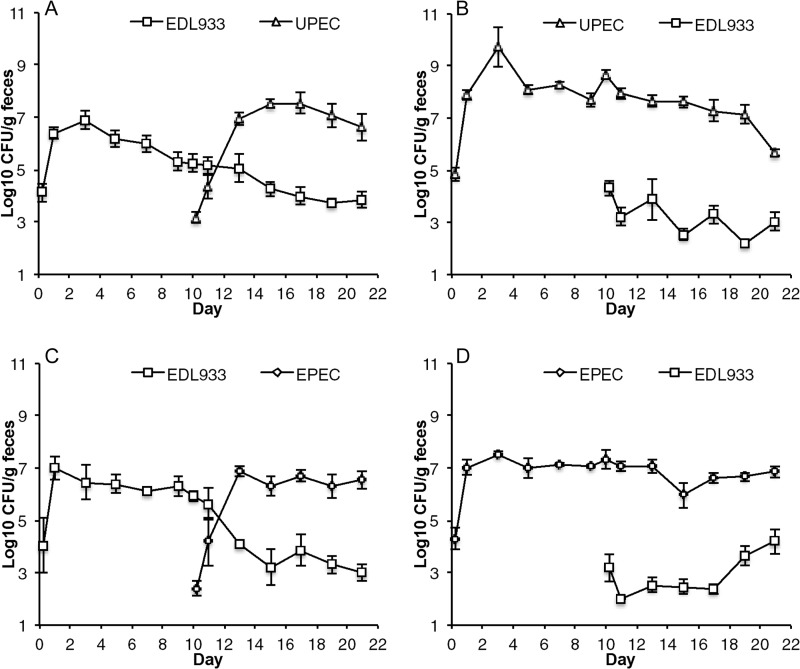
Since UPEC and EPEC can grow from low to high numbers in competition with precolonized E. coli HS and E. coli Nissle 1917 (Fig. 1) but E. coli EDL933 cannot (5), it can be inferred that UPEC and EPEC occupy intestinal niches that are distinct from that of EHEC. To test this possibility, we determined whether or not UPEC could overcome colonization resistance by the EHEC strain E. coli EDL933 (Fig. 3). Mice were precolonized with 105 CFU of EHEC by feeding and on day 10 challenged by association with 105 CFU of UPEC (Fig. 3A). UPEC overcame colonization resistance and grew to approximately 107 CFU per g of feces, suggesting that UPEC colonizes at least one niche that is not occupied by EHEC, whereas the population of EHEC declined from between 105 and 106 to approximately 104 CFU per g of feces for the duration of the 21-day experiment, suggesting that UPEC outcompetes EHEC in at least one niche. In a reciprocal experiment, mice were precolonized with UPEC and on day 10 challenged by association with 105 CFU of EHEC (Fig. 3B). UPEC colonized at between 107 and 108 CFU per g feces, whereas EHEC failed to grow further but persisted between 104 and 102 CFU per g of feces in competition with precolonized UPEC. Since in the absence of competition EHEC typically colonizes mice at approximately 107 CFU per g feces (10), these results indicate that UPEC outcompetes EHEC in its preferred niche(s).
FIG 3.
Competitive colonizations between E. coli pathotypes and an enterohemorrhagic strain. (A and B) Two sets of 3 mice were fed 105 CFU of E. coli EDL933 Strr Rifr and 10 days later were fed 105 CFU of E. coli CFT073 Strr Nalr (UPEC) (A), and the reciprocal experiment was conducted (B). (C and D) Two sets of 3 mice were fed 105 CFU of E. coli EDL933 Strr Rifr and 10 days later were fed 105 CFU of E. coli E2348/69 Strr Nalr (EPEC) (C), and the reciprocal experiment was conducted (D).
In analogous experiments, we determined whether or not EPEC and EHEC occupy distinct niches (Fig. 3). Mice were precolonized with 105 CFU of EHEC by feeding and on day 10 challenged by association with 105 CFU of EPEC (Fig. 3C). EPEC grew to approximately 107 CFU per g of feces, whereas the population of EHEC declined from approximately 106 to between 104 and 103 CFU per g of feces, suggesting that EPEC outcompetes EHEC in at least one niche. In a reciprocal experiment, mice were precolonized with EPEC and on day 10 challenged by association with 105 CFU of EHEC (Fig. 3D). EHEC failed to grow further but persisted between 104 and 102 CFU per g of feces in competition with precolonized EPEC, which colonized at approximately 107 CFU per g feces. These results corroborate the finding that EPEC outcompetes EHEC in its preferred niche(s). We conclude that UPEC and EPEC occupy intestinal niches that are not occupied by EHEC and apparently outcompete EHEC in its preferred niches.
UPEC and EPEC intestinal niches.
The data described above clearly demonstrate that UPEC and EPEC occupy intestinal niches that are distinct from those occupied by EHEC (Fig. 3) but do not distinguish whether or not their niches are the same or different. Therefore, we sought to determine if UPEC and EPEC occupy distinct intestinal niches by competing them in challenge experiments (Fig. 4). Mice were precolonized with 105 CFU of UPEC by feeding and on day 10 challenged by association with 105 CFU EPEC (Fig. 4A). EPEC failed to grow further but remained in the intestine at between 102 and 103 CFU per g of feces, whereas the population of UPEC remained at approximately 109 CFU per g of feces, suggesting that EPEC cannot compete effectively in any of the niches occupied by precolonized UPEC. In the reciprocal experiment, mice were precolonized by feeding 105 CFU of EPEC and on day 10 challenged by association with 105 CFU of UPEC (Fig. 4B). UPEC grew to approximately 106 CFU per g of feces, whereas the population of EPEC remained at approximately 109 CFU per g of feces, suggesting that UPEC occupies an additional niche that is not occupied by EPEC. In summary, UPEC and EPEC both occupy niches that are not occupied by EHEC (Fig. 3). UPEC occupies at least one niche that is not occupied by EPEC (Fig. 4B). However, EPEC does not appear to occupy any niches that are not occupied by UPEC (Fig. 4A). Finally, the niches shared by UPEC and EPEC are not occupied by commensal strains E. coli HS and E. coli Nissle 1917, as indicated by the inability of the commensals to prevent colonization by either UPEC or EPEC (Fig. 1).
FIG 4.
Competitive colonizations between UPEC and EPEC. (A) Two sets of 3 mice were fed 105 CFU of E. coli CFT073 Strr (UPEC) and 10 days later were fed 105 CFU of E. coli E2348/69 Strr Nalr (EPEC). (B) Two sets of 3 mice were fed 105 CFU of E. coli E2348/69 Strr (EPEC) and 10 days later were fed 105 CFU of E. coli CFT073 Strr Nalr (UPEC).
DISCUSSION
In this study, we showed that three different E. coli pathotypes occupy distinct niches in the streptomycin-treated mouse intestine (Fig. 3 and 4) and that two strains of the EHEC pathotype, E. coli EDL933 and E. coli Sakai, occupy the same niches in the intestine (Fig. 2). These findings have important implications for designing strategies to prevent colonization as a means to combat E. coli infections. Indeed, we demonstrated that a combination of commensal E. coli strains that previously was shown to prevent colonization by EHEC strain E. coli EDL933 (4) could not prevent colonization of UPEC or EPEC (Fig. 1). Therefore, it is unlikely that any particular commensal strain(s) of E. coli will be generally effective as a probiotic to prevent colonization by E. coli pathogens. Freter previously postulated that the specificity of microbial interactions in the intestine would pose a serious problem for selecting bacterial strains with optimal attributes as probiotics (30).
UPEC accounts for the majority of urinary tract infections in the United States (31), and these infections often are recurring, perhaps because the intestine serves as a reservoir for UPEC strains (32). Thus, it is reasonable to think that preventing these strains from colonizing the intestine would subsequently prevent infection of the urinary tract. Likewise, if a commensal strain could be found, or designed, to outcompete enteric pathogens (e.g., EHEC or EPEC) for limiting nutrients, it could potentially prevent E. coli infections, which account for up to 70% of infant diarrhea around the world (21). Thus, it is important to develop strategies to combat E. coli infections.
In humans, EPEC colonizes the small intestines and UPEC causes urinary tract infections (1). Since this is a study of human pathogens in mice, it is important to note that the streptomycin-treated mouse is a model for colonization, not for pathogenesis (4, 8). Subsequently, many aspects of human intestinal infections with E. coli, such as virulence factor expression and attachment, are not pertinent in mice. Indeed, it was established long ago that E. coli EDL933 does not cause an inflammatory response or elicit disease symptoms in streptomycin-treated mice (25). Here we show that when E. coli HS and E. coli Nissle 1917 are cocolonized with either E. coli E2348/69 (EPEC) or E. coli CFT073 (UPEC), both the EPEC and UPEC strains compete for the same intestinal sites as E. coli HS and Nissle 1917 (Table 2) and neither UPEC nor EPEC elicits obvious signs of inflammation in the streptomycin-treated mouse intestine.
According to Freter's nutrient niche hypothesis, an intestinal niche is defined by the nutrient(s) that supports growth of a bacterial population in the intestine (30). Metabolic modeling of nutrient utilization by 55 sequenced E. coli strains indicated modest differences, but the authors did not use the information to predict how the capacity to grow on certain nutrients might allow a strain to adapt to various environments (33). We have established previously that four different human commensal E. coli strains use the same nutrients in vitro (with one or two exceptions because the strain lacks the corresponding pathway), but each strain uses a subset of different nutrients in vivo, i.e., to colonize the streptomycin-treated mouse intestine (4, 5, 7–10, 14, 34). Colonized bacteria must effectively compete for the nutrients that allow them to grow faster than the turnover rate of the intestinal contents, or they will be washed out (3). This apparently includes bacteria that have the capacity to adhere to the mucosal epithelium, since we previously have shown that the majority of E. coli EDL933 cells are not tightly attached but rather reside in the loosely adherent mucus layer, where they can be sloughed into the intestinal contents (10). In this regard, colonization of the streptomycin-treated mouse intestine by E. coli follows the basic principles of Freter's nutrient niche hypothesis, which states that two species can coexist in the intestine only if each can grow better than all others on at least one nutrient (3).
We postulate three ways that E. coli strains compete for the nutrients that allow them to occupy distinct niches in the intestine. First, two strains can utilize different nutrients, as we have shown first for two competing E. coli strains (8) and more recently for four different strains (5). For the strains tested, each E. coli strain uses a complementary yet divergent set of seven sugars to colonize. Second, two strains can cocolonize, despite using the same set of sugars, by using some of those sugars better than one another, as we have shown for isogenic flhD mutants of E. coli K-12 that effectively outcompete their wild-type parent (12) by growing faster on some of the same sugars. Third, two strains can occupy different mixed biofilms (restaurants) in which different nutrients are made available by different polysaccharide-degrading anaerobes (14). Theoretically, these strategies for nutrient acquisition are not mutually exclusive, but how they intertwine to establish the intestinal niches occupied by each ecotype and pathotype is an open question.
Competitive colonizations between pathotypes suggest that at least some of them occupy more than one niche, even though the nature of those niches is not clear. When mice are precolonized with UPEC, EPEC is nearly eliminated when associated as a challenge strain (Fig. 4A), suggesting that UPEC competes effectively for all of the niches normally occupied by EPEC. On the other hand, in mice precolonized with EPEC, UPEC grows to a 10-fold-higher population (Fig. 4B), indicating that UPEC occupies one or more niches not occupied by EPEC. These results support the idea that the two strains compete for some of the same nutrients and/or restaurants but UPEC can use at least one of them more efficiently than EPEC.
Both UPEC and EPEC occupy niches that are not occupied by EHEC (Fig. 3). However, the two O157:H7 EHEC strains, E. coli EDL933 and E. coli Sakai, occupy the same niches (Fig. 2). These two strains are genetically nearly identical, having >99.9% sequence identity in their protein-coding genes (29) and relatively minor genomic differences that can be attributed to the way the two genomes were annotated (28), and thus it is likely that the two strains have very similar physiology, which is reflected by their occupation of the same niches in the intestine. The distinguishing features of the niches occupied by various E. coli ecotypes and pathotypes, as well as distinctive aspects of their physiology and metabolism that allows them to occupy these niches, are only partially understood. What is clear is that colonized E. coli, including invading pathogens, must compete effectively for the nutrients they need to grow and sustain their populations. These nutrients consist of mono- and disaccharides, as E. coli is primarily a consumer of simple sugars but cannot utilize the complex mucus- and fiber-derived polysaccharides that make up the bulk of carbon and energy sources in the intestine (8). It is the job of intestinal anaerobes to degrade complex polysaccharides, releasing the simple sugars that are used by other bacteria (15), including E. coli, to colonize.
Finally, we stress that colonization is the first step in enteric pathogenesis. Therefore, competition for the nutrients needed by pathogens to grow from low to high numbers in the intestine is essential for infection. A corollary of Freter's nutrient niche hypothesis is that limitation of nutrients can prevent invading strains from colonizing (30). Indeed, Freter found that E. coli C25 could prevent colonization by Shigella flexneri (35), and we showed that a mixture of commensal E. coli strains could prevent colonization by E. coli EDL933 (4, 5). But a note of caution is necessary. The average human is colonized with 5 commensal strains (36), yet frequent infections still occur, suggesting that many commensal E. coli ecotypes cannot prevent colonization by many E. coli pathotypes. Freter summarized the probiotic field by stating that there appears to exist a very high degree of specificity between antagonistic bacterial strains in the intestine (30). According to Freter's hypothesis, only strains that occupy the same niche will prevent invading pathogens from colonizing, i.e., only certain E. coli commensal strains will prevent colonization by other E. coli strains. Our study extends the idea that no single strain or combination of E. coli strains will prevent colonization by different E. coli pathotypes. While it might be possible to search for and find commensal E. coli strains that will exert colonization resistance against each E. coli pathotype, at this time we do not possess the knowledge needed to design a broad-spectrum probiotic strain, highlighting the need for a deeper understanding of the factors that contribute to the colonization of the mammalian gastrointestinal tract by E. coli.
ACKNOWLEDGMENTS
This research was supported by Public Health Service grant GM095370 to T.C. and P.S.C.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 2.Freter R, Brickner H, Botney M, Cleven D, Aranki A. 1983. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun. 39:676–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freter R. 1983. Mechanisms that control the microflora in the large intestine, p 33–54 In Hentges DJ. (ed), Human intestinal microflora in health and disease. Academic Press, Inc., New York, NY [Google Scholar]
- 4.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect. Immun. 77:2876–2886. 10.1128/IAI.00059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. 2013. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. 10.1371/journal.pone.0053957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS. 2007. L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 ΔfucAO and E. coli Nissle 1917 ΔfucAO mutants in the mouse intestine and in M9 minimal medium. Infect. Immun. 75:5465–5475. 10.1128/IAI.00822-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427–7432. 10.1073/pnas.0307888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76:1143–1152. 10.1128/IAI.01386-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, Struve C, Krogfelt KA, Cohen PS, Conway T. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 76:2531–2540. 10.1128/IAI.00096-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72:1666–1676. 10.1128/IAI.72.3.1666-1676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney NJ, Laux DC, Cohen PS. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabich AJ, Leatham MP, Grissom JE, Wiley G, Lai H, Najar F, Roe BA, Cohen PS, Conway T. 2011. Genotype and phenotypes of an intestine-adapted Escherichia coli K-12 mutant selected by animal passage for superior colonization. Infect. Immun. 79:2430–2439. 10.1128/IAI.01199-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leatham MP, Stevenson SJ, Gauger EJ, Krogfelt KA, Lins JJ, Haddock TL, Autieri SM, Conway T, Cohen PS. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039–8049. 10.1128/IAI.73.12.8039-8049.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leatham-Jensen MP, Frimodt-Moller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. 2012. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect. Immun. 80:1716–1727. 10.1128/IAI.06193-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nissle A. 1959. Explanations of the significance of colonic dysbacteria & the mechanism of action of E. coli therapy (mutaflor). Medizinische 4:1017–1022 [PubMed] [Google Scholar]
- 17.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 18.Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- 19.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787–796. 10.1093/oxfordjournals.aje.a010082 [DOI] [PubMed] [Google Scholar]
- 20.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conway T, Krogfelt KA, Cohen PS. 2007. Escherichia coli at the intestinal mucosal surface, p 175–196 In Brogden KA, Minion FC, Cornick N, Stanton TB, Zhang Q, Nolan LK, Wannemuehler MJ. (ed), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 23.Hentges DJ, Que JU, Casey SW, Stein AJ. 1984. The influence of streptomycin on colonization in mice. Microecol. Theor. 14:53–62 [Google Scholar]
- 24.Cohen PS, Laux DC. 1995. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 253:309–314. 10.1016/S0076-6879(95)53026-6 [DOI] [PubMed] [Google Scholar]
- 25.Wadolkowski EA, Burris JA, O'Brien AD. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukjancenko O, Wassenaar TM, Ussery DW. 2010. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60:708–720. 10.1007/s00248-010-9717-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook H, Ussery DW. 2013. Sigma factors in a thousand E. coli genomes. Environ. Microbiol. 15:3121–3129. 10.1111/1462-2920.12236 [DOI] [PubMed] [Google Scholar]
- 28.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. 2006. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 16:757–767. 10.1101/gr.4759706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freter R. 1992. Factors affecting the microecology of the gut, p 111–144 In Fuller R. (ed), Probiotics, the scientific basis. Chapman and Hall, London, United Kingdom [Google Scholar]
- 31.Welch RA, Burland V, Plunkett G, III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024. 10.1073/pnas.252529799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alteri CJ, Mobley HL. 2012. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 15:3–9. 10.1016/j.mib.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, Feist AM, Palsson BO. 2013. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl. Acad. Sci. U. S. A. 110:20338–20343. 10.1073/pnas.1307797110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Moller J, Krogfelt KA, Kazmierczak K, Kenney LJ, Conway T, Cohen PS, McCormick BA. 2014. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect. Immun. 82:670–682. 10.1128/IAI.01149-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freter R, Abrams GD. 1972. Function of various intestinal bacteria in converting germfree mice to the normal state. Infect. Immun. 6:119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apperloo-Renkema HZ, Van der Waaij BD, Van der Waaij D. 1990. Determination of colonization resistance of the digestive tract by biotyping of Enterobacteriaceae. Epidemiol. Infect. 105:355–361. 10.1017/S0950268800047944 [DOI] [PMC free article] [PubMed] [Google Scholar]