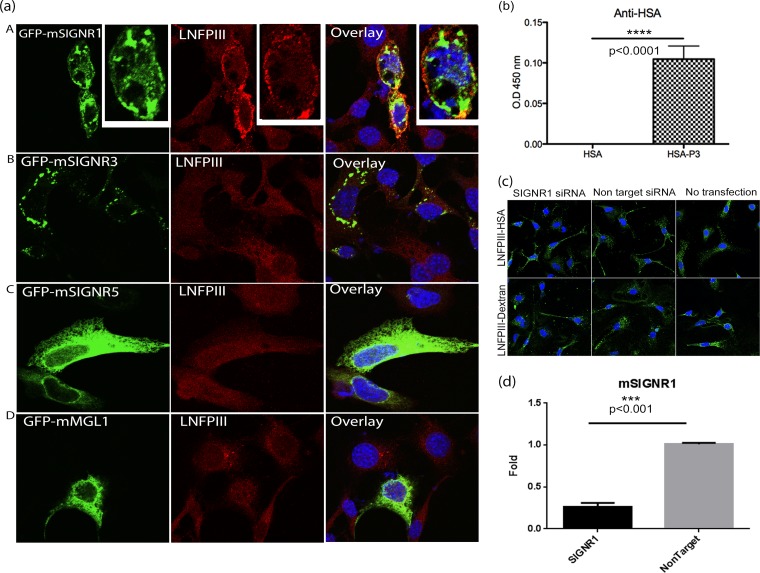
FIG 7.
LNFPIII-NGC binds to mouse SIGNR1 in vitro. (a) NIH 3T3 cells were transfected with green fluorescent protein (GFP)-fused constructs of mouse SIGNR1, SIGNR3, SIGNR5, or MGL (green). Cells were then incubated with LNFPIII-NGC (50 μg/ml), and endocytosis was induced for 30 min as described before. Cells were fixed, stained, and then analyzed under a confocal microscope. LNFPIII-NGCs are stained in red, SIGNRs in green, and overlay/colocalization in yellow. The above-described experiment represents 4 independent experiments repeated and reproduced 3 or 4 times. (b) LNFPIII-HSA directly binds to recombinant SIGNR1. ELISA was performed to confirm the direct interaction between SIGNR1 and LNFPIII-NGCs. Ninety-six-well plates were coated with recombinant SIGNR1 (10 μg/ml) overnight followed by incubation with LNFPIII-HSA conjugate (10 μg/ml) for 1 h at room temperature. Binding was detected using anti-HSA antibody followed by horseradish peroxidase (HRP)-conjugated secondary antibody. Recombinant HSA was used as a control. The data represent 4 independent experiment with n = 4. Statistical significance was calculated by Student's t test (****, P < 0.0001). O.D. 450 nm, optical density at 450 nm. (c) No change in LNFPIII-NGC uptake in macrophages lacking SIGNR1 expression. Endogenous SIGNR1 protein expression was downregulated in peritoneal macrophages by transfection with SIGNR1-specific siRNA for 36 h. Nonspecific siRNA was used as a control. Cells were then incubated with LNFPIII-NGC (50 μg/ml) for 30 min at 37°C as described previously. LNFPIII-NGC is stained in green. Data represent two individual experiments performed, giving similar results. (d) Validation of SIGNR1 protein expression knockdown by quantitative real-time PCR (qRT-PCR). Total RNAs from cells transfected with siRNA or nonspecific controls were used to measure expression of signr1 gene by the qRT-PCR method. Transfected samples were in triplicate, and the data shown are from 1 of 2 independent experiments. Statistical significance was calculated by Student's t test analysis (***, P < 0.001).