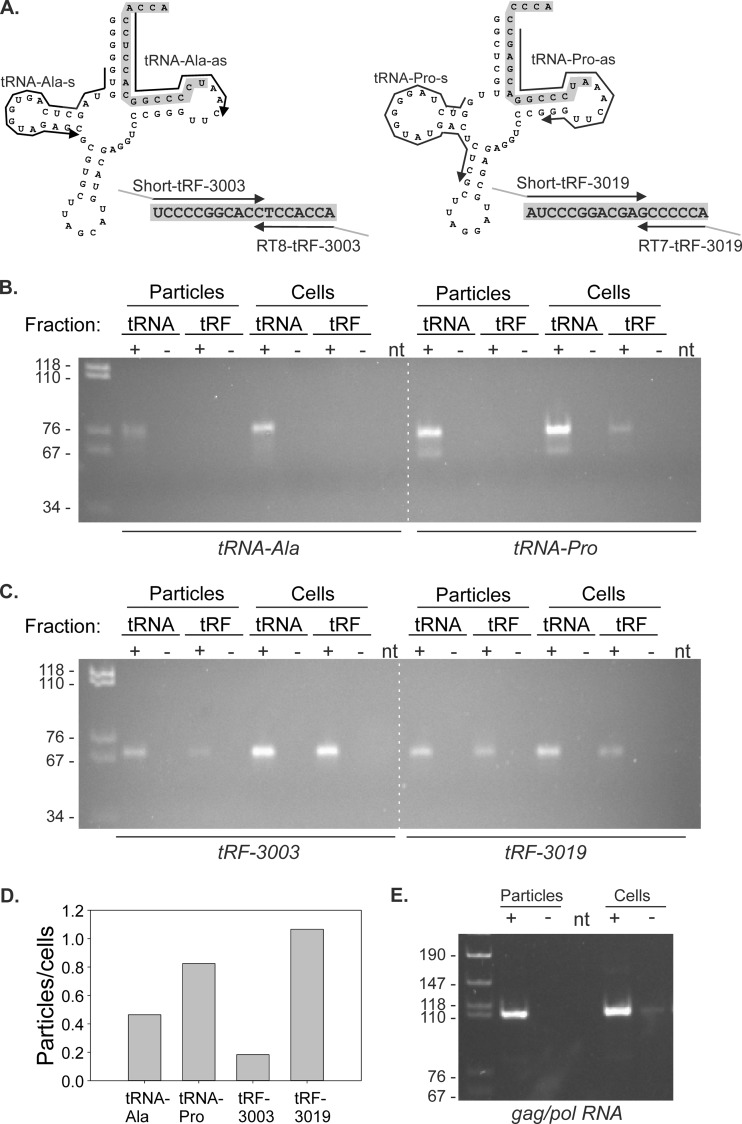
FIG 6.
RT-PCR to detect tRNAs, tRFs, and gag/pol RNA in virus particles and C91PL cells. (A) As described in Materials and Methods, RNA from virus particles and producer C91PL cells was subjected to denaturing PAGE; regions of the gel containing tRNA and tRFs were excised, and RNA was recovered by passive elution and ethanol precipitation. The resulting fractions were subjected to RT-PCR to detect tRNA-Ala, tRNA-Pro, and their tRF-3 sequences, tRF-3003 and tRF-3019, respectively. (B and C) Images of the RT-PCR products after separation on 6% polyacrylamide gels. The intensities of RT-PCR bands obtained for tRNAs and tRFs (measured in tRNA and tRF fractions, respectively) were measured using a Bio-Rad Gel Doc XRS imager. (D) Plot of ratios of band intensities obtained for virus particles versus cells. The calculated ratios were as follows: tRNA-Ala, particles/cells = 0.46; tRNA-Pro, particles/cells = 0.82; tRF-3003, particles/cells = 0.18; tRF-3019, particles/cells = 1.07. (E) Results of RT-PCR performed on RNA from the virus particles and producer cells to detect HTLV-1 genomic gag/pol RNA. RT-PCR was carried out using primers U5-s and Gag-as as described in Materials and Methods. The dashed white lines were added to panels B and C to aid in their alignment. The first lane on each gel contained MspI-digested pBluescript as a size marker; band sizes in basepairs are indicated on the left. The plus and minus signs above the lanes indicate RT reactions carried out in the presence (+) or absence (−) of reverse transcriptase. RNA template was omitted from the RT reaction in lanes labeled nt.