Abstract
The clinical management of meningitis caused by Escherichia coli is greatly complicated when the organism becomes resistant to broad-spectrum antibiotics. We sought to characterize the antimicrobial susceptibilities, sequence types (ST), and presence of known drug resistance genes of E. coli isolates that caused meningitis between 1996 and 2011 in Salvador, Brazil. We then compared these findings to those for E. coli isolates from community-acquired urinary tract infections (UTI) that occurred during the same time period and in the same city. We found that 19% of E. coli isolates from cases of meningitis and less than 1% of isolates from UTI were resistant to third-generation cephalosporins. The sequence types of E. coli isolates from cases of meningitis included ST131, ST69, ST405, and ST62, which were also found among isolates from UTI. Additionally, among the E. coli isolates that were resistant to third-generation cephalosporins, we found genes that encode the extended-spectrum beta-lactamases CTX-M-2, CTX-M-14, and CTX-M-15. These observations demonstrate that compared to E. coli strains isolated from cases of community-acquired UTI, those isolated from cases of meningitis are more resistant to third-generation cephalosporins, even though the same sequence types are shared between the two forms of extraintestinal infections.
INTRODUCTION
The prevalence of antimicrobial drug resistance in Gram-negative bacteria continues to increase worldwide, which complicates the clinical management of infections caused by these organisms (1). These infections include enteric as well as extraintestinal infections such as urinary tract infections (UTI), sepsis, and meningitis (2, 3). Escherichia coli is a Gram-negative bacillus that is capable of causing urinary tract infections (UTI) and meningitis, but it is also a normal constituent of the gut microbiota of mammals (2, 4).
E. coli isolates can be classified into pathomorphs based on the type of disease caused (2, 5). Johnson et al. termed E. coli strains isolated from extraintestinal sites as extraintestinal pathogenic E. coli (ExPEC) based on a distinct set of virulence genes (6). ExPEC can be further subdivided into pathomorphs that include those that cause urinary tract infection (uropathogenic E. coli [UPEC]) and those that cause neonatal meningitis, also known as neonatal meningitis-causing E. coli [NMEC] (2, 5, 7–9). This is an area of active research, but currently there are no virulence factors that unequivocally distinguish these groups of ExPEC.
It is thought that the E. coli strains that cause extraintestinal infections first colonize the gut (2). From the gut the E. coli organisms are then able to spread to the urinary tract and bloodstream (2). Animal models of E. coli meningitis have shown that once the E. coli organisms reach a high enough concentration in the bloodstream, they are able to cross the blood-brain barrier and cause meningitis (10, 11). The severity of meningitis has been correlated with the concentration of bacteria in the bloodstream (2). E. coli is the leading cause of UTI and second leading cause of neonatal meningitis in developed countries (2, 12). Although it is not as common, E. coli also causes meningitis in adults (12, 13).
Isolates of E. coli that have caused neonatal meningitis in developed countries have been extensively studied (10, 14–19). These studies have found that some serotypes and sequence types predominate among the E. coli strains that cause meningitis. However, these same serotypes and sequence types are found in E. coli strains isolated from other sources such as urinary tract infections, poultry, and healthy human digestive tracts (15, 16, 20). Serotypes of E. coli that have been isolated from cases of meningitis include O18:K1, O45:K1, O1:K1, and O83:K1 (14, 19, 21, 22). The sequence types isolated from cases of meningitis include ST95, ST62, ST131, and ST69 (14, 19, 20, 22–24). When combinations of typing methods and more discriminant typing methods are used, isolates that have caused meningitis can be separated from isolates from other sources (15, 16). However, most studies examined a limited number of collections of E. coli strains available for study obtained from different geographic sites and time periods, and, hence, it is unclear how significant these differences are.
There are recent reports of meningitis caused by E. coli strains that are resistant to third-generation cephalosporins and produce extended-spectrum beta-lactamases (ESBL) (23, 25). The spread of E. coli strains that are resistant to broader-spectrum cephalosporin antibiotics is problematic because these antibiotics are recommended for the treatment of meningitis caused by E. coli (11, 12). We sought to characterize E. coli strains isolated from UTI and meningitis patients by both multilocus sequencing typing (MLST) and the presence of ESBL genes in order to better understand the spread and distribution of drug resistance in E. coli strains that cause extraintestinal infections.
MATERIALS AND METHODS
Isolate collection.
The isolates characterized in this study were originally collected as part of three separate studies of meningitis and community-acquired UTI that occurred in Salvador, Brazil (26, 27). Due to sample loss, only a subset of the UTI isolates were characterized. All E. coli isolates from the meningitis collection were included in the current study.
As part of active hospital-based surveillance, E. coli isolates from cases of meningitis that occurred between 1996 and 2011 in Salvador were consecutively collected. The study was based at Courto Maia Hospital, which serves as the infectious disease referral hospital and reference laboratory. This laboratory is part of the public health system, and all cases of meningitis that occur in the region are required to be reported to this laboratory. As part of an ongoing study of the etiology of meningitis, limited epidemiologic information is recorded, and bacterial isolates are collected from this reference laboratory and then biobanked at the Gonçalo Moniz Research Center. The methodology and results of this study were previously published (27).
E. coli isolates from community-acquired UTI were collected as part of two separate studies, one that occurred between 2001 and 2002 and another that occurred between 2008 and 2009. The results and methodology of the study of E. coli isolates from UTI that occurred between 2001 and 2002 have been published elsewhere (26). The study of E. coli isolates collected from UTI from 2008 to 2009 is unpublished (M. G. Barberino, J. N. Reis, and E. D. Moreira, Jr., personal communication). The antimicrobial susceptibilities of the UTI E. coli isolates reported here were determined as part of this previous study. The 2008-2009 UTI study by Barberino et al. occurred at a private outpatient emergency room in Salvador, Brazil, and was conducted in accordance with all relevant protections for human subjects (institutional review board [IRB] 180/2008).
As previously published, the isolates from the 2001-2002 and the 2008-2009 UTI collections were classified as susceptible or nonsusceptible to each antimicrobial agent tested. Nonsusceptible isolates included isolates that showed either intermediate or resistant phenotypes.
Antimicrobial susceptibility testing.
The susceptibilities of the E. coli isolates from cases of meningitis were determined with MicroScan BP38 panels (Siemens). ATCC 27853 and ATCC 25922 strains were used as reference strains for all tests.
According to the manufacturers' published breakpoints, isolates were classified as susceptible, intermediate, or resistant to each drug. Where indicated, isolates were classified as either susceptible or nonsusceptible. The nonsusceptible isolates contained all those with either the intermediate or resistant phenotype.
DNA extraction, beta-lactamase detection, and sequencing.
DNA was extracted with a Promega Wizard genomic DNA kit according to the manufacturer's recommendations.
Meningitis isolates were screened for the presence of blaTEM, blaSHV, blaOXA, and blaCTX-M gene sequences with multiplex PCR protocols previously published by Dallenne et al. (28). UTI isolates were screened for only blaTEM, blaSHV, and blaOXA. The primer sets used were Multiplex I TEM, SHV, and OXA-1-like and Multiplex II CTX-M group 1, group 2, and group 9 (28). All reactions and gel electrophoresis were performed according to previously published protocols (28). For each reaction, DNA known to contain the relevant gene sequence was used as a positive control, and water was used as a negative control.
The PCR products that tested positive for a beta-lactamase were submitted for sequencing to the University of California, Berkeley, DNA Sequencing Facility. The resulting sequences were then submitted to BLASTN for identification (29).
Multilocus sequence typing and sequence analysis.
Multilocus sequence typing (MLST) was performed according to previously published protocols (20). The sequences were submitted to the online MLST database at http://mlst.ucc.ie/mlst/dbs/Ecoli for sequence type (ST) identification.
All sequencing was done at the University of California, Berkeley, DNA Sequencing Facility. The resulting sequences were cleaned, assembled, and analyzed with Geneious 5.6.
Data analysis.
All data analyses and graphs were generated with STATA 10. P values were calculated with Fisher's exact test.
RESULTS
Characterization of E. coli causing meningitis.
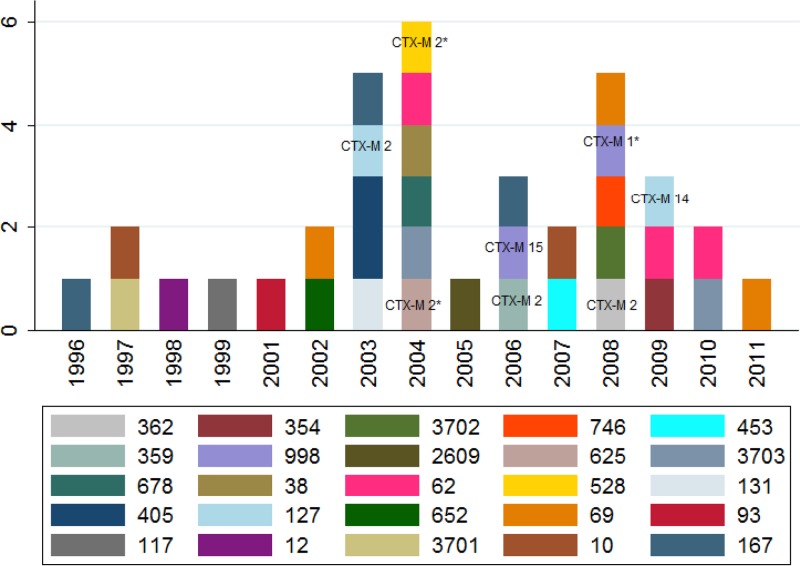
Thirty-six E. coli isolates from cases of meningitis were collected from patients in Salvador, Brazil, from 1996 to 2010. Figure 1 displays the year of isolation and sequence type of each of these isolates. The ages of the patients who had meningitis were known for 21 isolates. Thirteen (62%) of these isolates were from children <2 years of age. Three novel sequence types (ST3701, ST3702, and ST3703) were identified among these isolates. All of the isolates with novel sequence types were isolated from cases of meningitis in children who were <2 years old.
FIG 1.
Sequence types versus year of isolate collection. Isolates that were resistant to third-generation cephalosporins are labeled with the blaCTX-M-type gene they carry. Isolates marked with an asterisk did not yield an interpretable DNA sequence and are labeled with the name of the blaCTX-M group they tested positive for.
Among the meningitis isolates, 19% were resistant to ceftazidime and cefotaxime, 17% were resistant to aztreonam, and 14% were resistant to ciprofloxacin (Table 1). All isolates that were resistant to third-generation cephalosporins were found to have caused meningitis after 2002 (Fig. 1). We did not find resistance to carbapenems, cefotetan, or tigecycline.
TABLE 1.
Results of antimicrobial susceptibility testing of 36 E. coli isolates from cases of meningitisa
| Antimicrobial |
E. coli isolates that were: |
|||||
|---|---|---|---|---|---|---|
| Susceptible |
Intermediate |
Resistant |
||||
| No. | % | No. | % | No. | % | |
| Amikacin | 32 | 89 | 1 | 4 | 3 | 8 |
| Gentamicin | 31 | 86 | NA | NA | 5 | 14 |
| Tobramycin | 31 | 86 | NA | NA | 5 | 14 |
| Chloramphenicol | 27 | 75 | 1 | 4 | 8 | 22 |
| Tetracycline | 15 | 42 | NA | NA | 21 | 58 |
| Nitrofurantoin | 35 | 97 | NA | NA | 1 | 3 |
| Ciprofloxacin | 31 | 86 | NA | NA | 5 | 14 |
| Levofloxacin | 32 | 89 | NA | NA | 4 | 11 |
| Piperacillin | 13 | 36 | 1 | 4 | 22 | 61 |
| Ampicillin-sulbactam | 17 | 47 | 5 | 19 | 14 | 39 |
| Piperacillin-tazobactam | 33 | 92 | 2 | 8 | 1 | 3 |
| Aztreonam | 30 | 83 | NA | NA | 6 | 17 |
| Cefazolin | 25 | 69 | 1 | 4 | 10 | 28 |
| Cefepime | 31 | 86 | NA | NA | 5 | 14 |
| Cefotaxime | 28 | 78 | 1 | 4 | 7 | 19 |
| Cefoxitin | 35 | 97 | 1 | 4 | 0 | 0 |
| Ceftazidime | 29 | 81 | NA | NA | 7 | 19 |
| Ceftriaxone | 28 | 78 | 1 | 4 | 7 | 19 |
| Cefuroxime | 28 | 78 | NA | NA | 8 | 22 |
| Cephalothin | 12 | 33 | 11 | 42 | 13 | 36 |
| Trimethoprim-sulfamethoxazole | 12 | 33 | NA | NA | 24 | 67 |
The categories marked with NA were not tested. Beta-lactam and beta-lactamase inhibitor combinations are ampicillin-sulbactam and piperacillin-tazobactam. Aztreonam is a monobactam. First-generation cephalosporins are cephalothin and cefazolin, second-generation cephalosporins include cefoxitin and cefuroxime, third-generation cephalosporins are cefotaxime and ceftazidime, and the fourth-generation cephalosporin is cefepime.
Eighty-six percent of isolates carried blaTEM, 13% carried blaSHV, and none carried blaOXA. Thirty-six isolates tested positive for blaCTX-M. All isolates that were resistant to third-generation cephalosporins carried a blaCTX-M-type gene. Five percent of isolates tested positive for blaCTX-M group 1, 3% for blaCTX-M group 9, and 28% for blaCTX-M group 2. Of the 13 isolates that tested positive for CTX-M, 10 yielded interpretable sequences. blaCTX-M −2 was identified in isolates ST652, ST62, ST405, ST127, ST167, ST359, and ST362. blaCTX-M-15 was found in one ST998 isolate. blaCTX-M-14 was found in one ST127 isolate (Fig. 1).
Comparison of E. coli isolates that caused UTI and meningitis.
The E. coli strains isolated from cases of meningitis had a significantly (P < 0.001) higher frequency of resistance to third-generation cephalosporins than the E. coli strains that were isolated from UTI. The study of the 544 cases of UTI in Salvador that took place in 2001-2002 found that <1% of isolates were nonsusceptible (intermediate or resistant phenotypes) to third-generation cephalosporins (26). The study of the 411 cases of community-acquired UTI that took place between 2008 and 2009 found that all isolates were susceptible to third-generation cephalosporins. The current study of the 36 E. coli isolates from cases of meningitis found that 22% were nonsusceptible to third-generation cephalosporins and that 19% were resistant to third-generation cephalosporins.
We compared the E. coli strains that caused both UTI and meningitis by comparing the sequence types of isolates with identical drug-resistant phenotypes. The isolates were classified as susceptible or nonsusceptible to eight classes of antimicrobial drugs because different antimicrobial susceptibility testing methods, drugs, and breakpoints were used to characterize each collection of E. coli isolates. Only the classes of drugs that all isolates were tested with were included in this analysis. These classes were tetracyclines (TET), aminoglycosides (AMI), trimethoprim-sulfamethoxazole (TSX), penicillins (PEN), first-generation cephalosporins (CEPH1), third generation cephalosporins (CEPH3), fluoroquinolones (FLR), and nitrofurantoin (NIT). Each drug resistance phenotype was named after the classes of drugs for which the isolates were nonsusceptible.
The drug resistance phenotypes found in both the UTI and meningitis collections were TET+ TSX+ PEN+ CEPH1, TSX+ PEN+ CEPH1, TET+ TSX+ PEN, PEN+ CEPH1, TET+ TSX+ CEPH1, TET+ TSX, TET+ TSX+ FLR, and TET+ TSX+ FLR+ CEPH1 (Table 2). The available UTI isolates with these phenotypes were submitted for multilocus sequence typing and screened for the blaTEM, blaSHV, and blaOXA gene sequences. From these isolates, 10 sequence types were identified in both the meningitis and UTI collections. These sequence types were ST10, ST62, ST69, ST93, ST127, ST131, ST167, ST405, ST998, and ST2609.
TABLE 2.
Drug-resistant phenotypes, beta-lactamase (blaTEM, blaSHV, or blaOXA), and sequence types found in collections of E. coli from meningitis and UTIa
| Phenotype | Results for meningitis |
Results for UTI in: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 2001-2002 |
2008-2009 |
||||||||
| No. | Sequence type(s) | bla gene(s) | No. | Sequence type(s) | bla | No. | Sequence type(s) | bla gene(s) | |
| TET+ TSX+ PEN+ CEPH1 | 5 | 167, 678, 69 | blaTEM | 6 | 394, 127, 998, 38, 73 | blaTEM | 18 | 38, 69, 93, 104, 31, 998, 14, 131, 73, 405, 62, 372, 3,928 | blaTEM, blaSHV |
| TSX+ PEN+ CEPH1 | 4 | 3703, 2609, 62 | blaTEM | 4 | 62, 38, 73, 127 | blaTEM | 15 | 69, 62, 10, 12, 14, 23, 73, 38, 12, 2.609 | blaTEM |
| TET+ TSX+ PEN | 4 | 69, 746, 3702 | blaTEM | 24 | 550, 167, 976, 3,704, 73, 69, 131, 3.910, 1,312, 3,704, 205, 38, 10 | blaTEM | 7 | 69, 95, 14, 38, 74, 827 | blaTEM, blaSHV |
| PEN+ CEPH1 | 2 | 652, 62 | blaTEM | 3 | 127, 10, 73 | blaTEM | 6 | 3,705, 73, 3,706, 38, 127 | blaTEM |
| TET+ TSX+ CEPH1 | 2 | 405 | blaTEM | 1 | 46 | blaTEM | None | None | None |
| TET+ TSX | 1 | 10 | blaTEM, blaSHV | 10 | 10, 3903, 167, 73, 127, 46, 3,904, 12, 998 | blaTEM | 1 | 394 | None |
| TET+ TSX+ FLR | 1 | 117 | blaTEM, blaSHV | 1 | 533 | None | 0 | None | None |
| TET+ TSX+ FLR+ CEPH1 | 1 | 354 | None | 0 | None | None | 1 | 3.902 | blaSHV |
UTI, urinary tract infection.
Of these common sequence types, three types (ST10, ST62, and ST69) had the same drug resistance phenotypes in both the meningitis and UTI collections. Of these three sequence types, only ST69 and ST62 showed the same beta-lactamase genotype in the UTI and meningitis collections.
Isolates with the phenotype TET+ SXT+ PEN+ CEPH1 that were ST69 and that carried the blaTEM gene were isolated from a case of meningitis in 2008 and from six cases of UTI from 2008 to 2009. Isolates with the phenotype TET+ SXT+ PEN that were ST69 and that carried the blaTEM gene were isolated from a case of meningitis in 2001, six cases of UTI that occurred between 2001 and 2002, and one case of UTI that occurred between 2008 and 2009. Isolates with the phenotype SXT+ PEN+ CEPH1 that were ST62 and that carried the blaTEM gene were isolated from two cases of meningitis in 2009 and 2010. ST62 with blaTEM and the same drug resistance phenotype was also isolated from one UTI that occurred between 2001 and 2002 and from one UTI that occurred between 2008 and 2009.
DISCUSSION
To better understand the spread of drug-resistant E. coli strains, we compared MLST and presence of blaTEM and blaSHV beta-lactamase genes from E. coli strains isolated from UTI and meningitis cases. We strain typed the E. coli by an MLST protocol which is commonly used to characterize drug-resistant E. coli isolates (20, 30). We identified a number of internationally distributed sequence types that are frequently drug resistant. These sequence types have also previously been isolated from food products and include the ST10 complex, ST69, ST117, ST131, and ST405 (30). ST69, previously labeled CgA, was first reported from UTI patients in Berkeley, California, USA, and has also been reported from UTI patients in Rio de Janeiro, Brazil (31, 32). ST62 has previously been associated with K1 E. coli, a serotype that frequently causes neonatal meningitis (20). Our findings further highlight the worldwide distribution of these lineages and the fact that they are capable of causing meningitis.
The results of antimicrobial susceptibility testing of the E. coli strains that caused meningitis showed that 19% were resistant to third-generation cephalosporins. These drugs are currently recommended for the treatment of meningitis suspected to be caused by Gram-negative organisms (12). In this study, we also identified extended-spectrum beta-lactamase genes in all meningitis isolates that were resistant to third-generation cephalosporins. In addition to the presence of blaTEM and blaSHV genes, this study identified blaCTX-M-2, blaCTX-M-15, and blaCTX-M-14 genes among the E. coli strains that caused meningitis. Cases of meningitis caused by ESBL-producing E. coli strains have been also been reported from Turkey, France, Algeria, Thailand, Germany, and Brazil (23, 33).
The E. coli strains that caused meningitis were more resistant to third-generation cephalosporins than those that caused community-acquired UTI. While the numbers of E. coli strains that caused meningitis are small (n = 36), this difference was still statistically significant. Unfortunately, the UTI isolates that were nonsusceptible to third-generation cephalosporins were lost, and we were unable to investigate these isolates further.
The results of this study support the findings of previous studies comparing E. coli strains from cases of meningitis and E. coli strains isolated from other sources (14, 15, 21). These studies showed that the same sequence types are frequently found in E. coli strains that are isolated from meningitis and other sources, and once further typing schemes are applied, differences can be found. In our study, these overlapping sequence types included the most common E. coli sequence types in the UTI collection. When the drug resistance profiles and beta-lactamase genotypes were compared, many of the E. coli strains from the meningitis cases could be further differentiated from those that caused UTI. The meningitis-causing E. coli strains may express additional factors that render them more invasive.
ACKNOWLEDGMENTS
We acknowledge the team at the Gonçalo Moniz Research Center, who coordinated the meningitis study and assisted with the laboratory work. This team included Jailton Azevedo Silva, Maira Santos, and Lorena Freire.
This project was supported in part by the Fogarty International Center and the National Institute of Allergy and Infectious Diseases under grant R25TW009338.
Footnotes
Published ahead of print 12 February 2014
REFERENCES
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 3.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26–38. 10.1038/nrmicro2265 [DOI] [PubMed] [Google Scholar]
- 4.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 5.Russo T, Johnson J. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754. 10.1086/315418 [DOI] [PubMed] [Google Scholar]
- 6.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161–2168. 10.1128/AAC.47.7.2161-2168.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson J, Russo T. 2005. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 295:383–404. 10.1016/j.ijmm.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 8.Johnson T, Wannemuehler Y, Johnson S, Stell A, Doetkott C, Johnson J, Kim K, Spanjaard L, Nolan L. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043–7050. 10.1128/AEM.01395-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foxman B. 2010. The epidemiology of urinary tract infection. Nat. Rev. Urol. 7:653–660. 10.1038/nrurol.2010.190 [DOI] [PubMed] [Google Scholar]
- 10.Kim KS. 2003. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat. Rev. Neurosci. 4:376–385. 10.1038/nrn1103 [DOI] [PubMed] [Google Scholar]
- 11.Kim KS. 2012. Current concepts on the pathogenesis of Escherichia coli meningitis: implications for therapy and prevention. Curr. Opin. Infect. Dis. 25:273–278. 10.1097/QCO.0b013e3283521eb0 [DOI] [PubMed] [Google Scholar]
- 12.Kim KS. 2010. Acute bacterial meningitis in infants and children. Lancet Infect. Dis. 10:32. 10.1016/S1473-3099(09)70306-8 [DOI] [PubMed] [Google Scholar]
- 13.Choi C. 2001. Bacterial meningitis in aging adults. Clin. Infect. Dis. 33:1380–1385. 10.1086/322688 [DOI] [PubMed] [Google Scholar]
- 14.Bidet P, Mahjoub-Messai F, Blanco J, Blanco J, Dehem M, Aujard Y, Bingen E, Bonacorsi S. 2007. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J. Infect. Dis. 196:297–303. 10.1086/518897 [DOI] [PubMed] [Google Scholar]
- 15.Logue CM, Doetkott C, Mangiamele P, Wannemuehler YM, Johnson TJ, Tivendale KA, Li G, Sherwood JS, Nolan LK. 2012. Genotypic and phenotypic traits that distinguish neonatal meningitis-associated Escherichia coli from fecal E. coli isolates of healthy human hosts. Appl. Environ. Microbiol. 78:5824–5830. 10.1128/AEM.07869-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78:3412–3419. 10.1128/IAI.00347-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houdouin V, Bonacorsi S, Bidet P, Bingen E. 2007. Antimicrobial susceptibility of 136 Escherichia coli isolates from cases of neonatal meningitis and relationship with virulence. Clin. Microbiol. Infect. 13:1207–1210. 10.1111/j.1469-0691.2007.01838.x [DOI] [PubMed] [Google Scholar]
- 18.Houdouin V, Bonacorsi S, Bidet P, Blanco J, De La Rocque F, Cohen R, Aujard Y, Bingen E. 2008. Association between mortality of Escherichia coli meningitis in young infants and non-virulent clonal groups of strains. Clin. Microbiol. Infect. 14:685–690. 10.1111/j.1469-0691.2008.02019.x [DOI] [PubMed] [Google Scholar]
- 19.Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Medigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun. 77:2272–2284. 10.1128/IAI.01333-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson JR, Delavari P, O'Bryan TT. 2001. Escherichia coli O18:K1:H7 isolates from patients with acute cystitis and neonatal meningitis exhibit common phylogenetic origins and virulence factor profiles. J. Infect. Dis. 183:425–434. 10.1086/318086 [DOI] [PubMed] [Google Scholar]
- 22.Bonacorsi S, Bingen E. 2005. Molecular epidemiology of Escherichia coli causing neonatal meningitis. Int. J. Med. Microbiol. 295:373–381. 10.1016/j.ijmm.2005.07.011 [DOI] [PubMed] [Google Scholar]
- 23.Elaldi N, Gozel MG, Kolayli F, Engin A, Celik C, Bakici MZ, Vahaboglu H. 2013. Community-acquired CTX-M-15-type ESBL-producing Escherichia coli meningitis: a case report and literature review. J. Infect. Dev. Ctries. 7:424–431. 10.3855/jidc.2820 [DOI] [PubMed] [Google Scholar]
- 24.Andrade LN, Minarini LA, Pitondo-Silva A, Climaco EC, Palazzo IC, Medeiros MI, Darini AL. 2010. Determinants of β-lactam resistance in meningitis-causing Enterobacteriaceae in Brazil. Can. J. Microbiol. 56:399–407. 10.1139/W10-020 [DOI] [PubMed] [Google Scholar]
- 25.Chaiyakulsil C, Prommalikit O. 2012. Successful medical treatment in a child with E. coli ESBL meningitis with acute communicating hydrocephalus and ventricular empyema: a case report. J. Med. Assoc. Thai. 95(Suppl 12):S138–S141 [PubMed] [Google Scholar]
- 26.Moreira ED, Jr, De Siqueira IC, Alcantara AP, Guereiro De Moura CG, De Carvalho WA, Riley L. 2006. Antimicrobial resistance of Escherichia coli strains causing community-acquired urinary tract infections among insured and uninsured populations in a large urban center. J. Chemother. 18:255–260. 10.1179/joc.2006.18.3.255 [DOI] [PubMed] [Google Scholar]
- 27.Lima JBT, Ribeiro GS, Cordeiro SM, Gouveia EL, Salgado K, Spratt BG, Godoy D, Reis MG, Ko AI, Reis JN. 2010. Poor clinical outcome for meningitis caused by Haemophilus influenzae serotype a strains containing the iS1016-bexA deletion. J. Infect. Dis. 202:1577–1584. 10.1086/656778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 29.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden T. 2008. NCBI BLAST web site NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9. 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manges AR, Johnson JR. 2012. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 55:712–719. 10.1093/cid/cis502 [DOI] [PubMed] [Google Scholar]
- 31.Dias RC, Marangoni DV, Smith SP, Alves EM, Pellegrino FL, Riley LW, Moreira BM. 2009. Clonal composition of Escherichia coli causing community-acquired urinary tract infections in the State of Rio de Janeiro, Brazil. Microb. Drug Resist. 15:303–308. 10.1089/mdr.2009.0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007–1013. 10.1056/NEJMoa011265 [DOI] [PubMed] [Google Scholar]
- 33.Heideking M, Lander F, Hufnagel M, Pfeifer Y, Wicker E, Krause G, Berner R. 2013. Antibiotic susceptibility profiles of neonatal invasive isolates of Escherichia coli from a 2-year nationwide surveillance study in Germany, 2009-2010. Eur. J. Clin. Microbiol. Infect. Dis. 32:1221–1223. 10.1007/s10096-013-1871-3 [DOI] [PubMed] [Google Scholar]