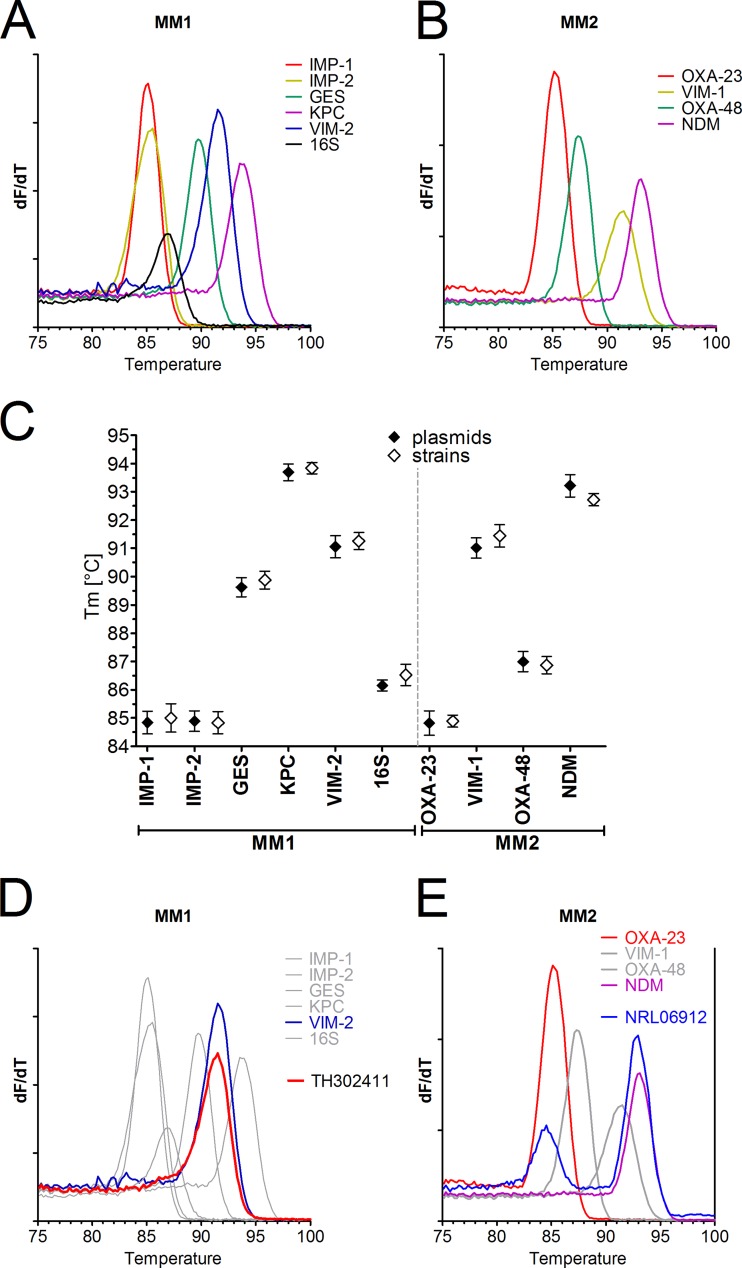
FIG 1.
Development of a multiplex PCR for detection of carbapenemases based on amplicon melting point differences. Plasmids encompassing amplicons of the indicated carbapenemases were amplified by a SYBR green multiplex PCR mix for IMP-1, IMP-2 (IMP-8/-16), GES, VIM-2, KPC, and 16S rRNA (master mix 1 [MM1]) (A) or OXA-23-like, OXA-48-like, VIM-1, and NDM (master mix 2 [MM2]) in the PCR-only mode on the BD Max cycler followed by melt curve analysis (B). The first derivate dF/dT is shown. (C) Plasmids were tested in 4 replicates on 3 different BD Max cyclers (n = 12, mean ± SD) (closed symbols). In a similar manner, one typical strain from a reference collection, encoding a respective carbapenemase, was tested in 3 replicates on 3 BD Max cyclers (n = 9, mean ± SD) (open symbols). The melting point temperatures (Tm) are indicated for the various amplicons detected by MM1 and MM2. (D) A clinical isolate (TH302411) analyzed by amplification with master mix 1 was identified as VIM-2 positive by comparison to the reference panel. (E) An isolate (NRL06912) with two carbapenemases (OXA-23 and NDM) was tested with master mix 2.