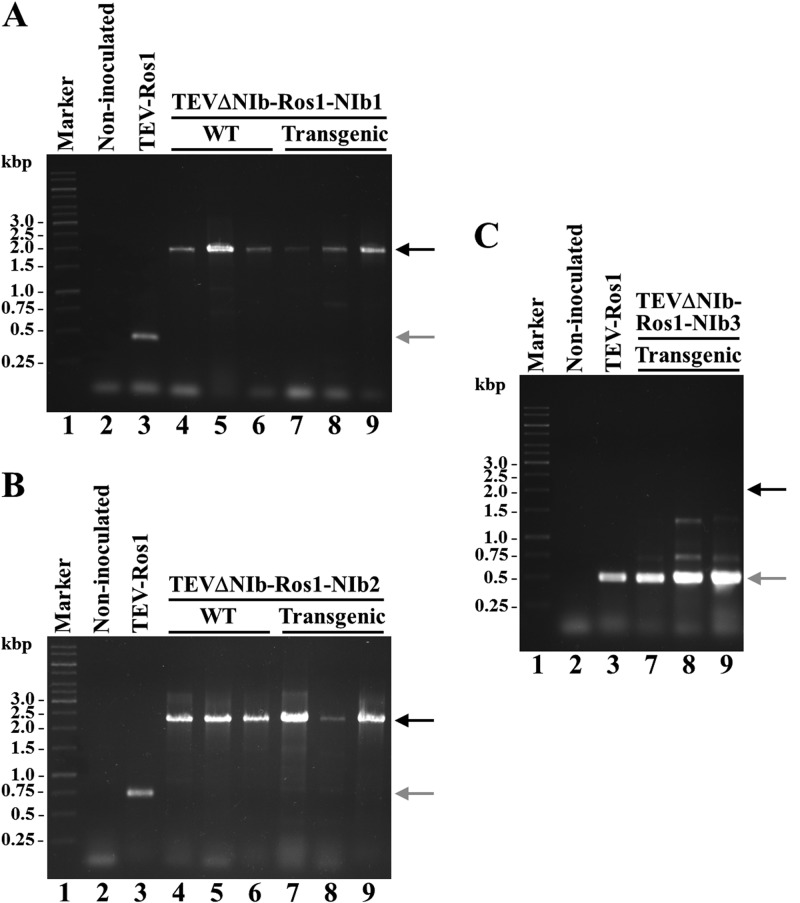
FIG 3.
Stability of the relocated NIb gene in the viral progeny at 15 d.p.i. RNA from systemic leaves of three representative inoculated plants was purified. cDNAs amplified by RT-PCR using primers flanking the new NIb location for TEVΔNIb-Ros1-NIb1 (A), TEVΔNIb-Ros1-NIb2 (B), and TEVΔNIb-Ros1-NIb3 (C) were separated by electrophoresis in 1% agarose gels, and stained with ethidium bromide. Lanes 1, DNA marker with the sizes of some of the components indicated on the left; lanes 2, noninoculated negative control; lanes 3, parental virus positive control (TEV-Ros1); lanes 4 to 6, three infected wild-type plants; lanes 7 to 9, three infected transgenic plants. Note that in panel C, lanes 4 to 6 (wild-type plants) are missing because none of these plants was infected. The black and gray arrows indicate the positions of the expected RT-PCR products if the relocated NIb is present or absent, respectively. Primers used to amplify the different cDNAs were, for RT reactions, 5′-CTTTACATAGTTTTTTTCCAACATTTCATG-3′, and for PCRs, 5′-AAAATAACAAATCTCAACACAACATATAC-3′ and 5′-CTCTTGCCATGGGTGAGCGCGCGAC-3′ (A), 5′-CTCAACTCCAAGAATTTC-3′ and 5′-GTCGCGCGCTCACCCATGGCAAGAG-3′ (B), and 5′-GTGGCCAACAATGCAAGATGTTGC-3′ and 5′-GTTAACTTTTGCGCCAAGGCTGAC-3′ (C).