FIG 2.
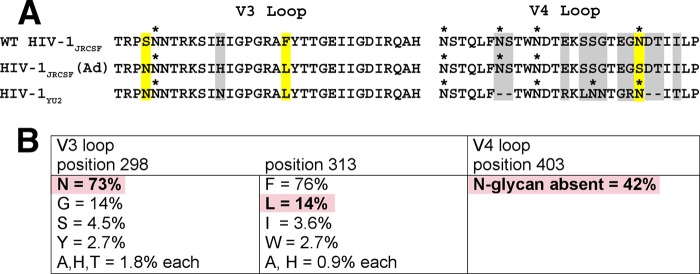
Adaptive mutations present in HIV-1JRCSF(Ad) occur commonly in many HIV-1 isolates. (A) Protein sequence alignment of V3 and V4 loops from wild-type HIV-1JRCSF, HIV-1JRCSF(Ad), and HIV-1YU2. HIV-1YU2 has V3 loop 298N and 313L residues that are also present in HIV-1JRCSF(Ad) but, unlike HIV-1JRCSF(Ad), possesses an N-glycan at position 403 within the heavily glycosylated V4 loop. Adaptive mutations are outlined in yellow, and glycosylation sites are marked with an asterisk. Other HIV-1YU2 residues that are different from those in HIV-1JRCSF(Ad) but are not known to contribute to entry activation by Nt are outlined in gray. The HXB2 standard numbering for HIV-1JRCSF(Ad) adaptive mutations is 300N, 317L, and 411S. The alignment was generated using MacVector software (version 7.2.3). (B) HIV-1JRCSF(Ad) adaptive mutations occur commonly in natural HIV-1 isolates. The frequencies of adaptive mutations in V3 and V4 loop sequences from 112 HIV-1 isolates, including isolates of clades A to D, F to H, J, and K, were analyzed using sequence data from the HIV Sequence Compendium 2013. Three clade D isolates (D.CM.10.DEMD10CM009, D.KE.97.ML415 2, and D.TD.99.MN011) and one clade B isolate (B.ES.09.P2149 3), representing 3.6% of total isolates analyzed, possessed all three mutations. Due to the hypervariability of the V4 loop, the location of position 403 within diverse isolates relative to its location within HIV-1JRCSF is approximate. Additionally, approximately 6% of the HIV-1 isolates surveyed did not possess an N-glycan within 2 amino acids of position 403. Pink boxes, adaptive mutations present in HIV-1JRCSF(Ad) that also occur in diverse HIV-1 isolates.
