FIG 1.
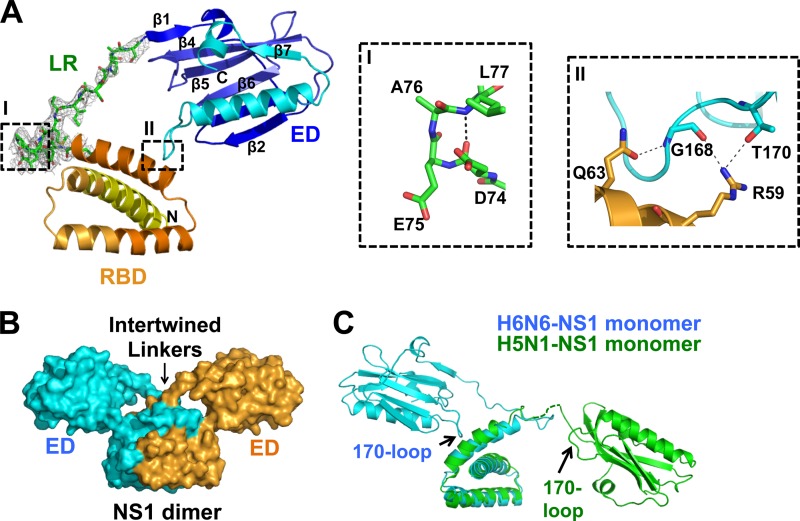
H6N6-NS1 structure. (A) Diagram representation of H6N6-NS1 structure with the RNA-binding domain (RBD) (yellow to orange), linker region (LR) (green), and effector domain (ED) (blue to cyan). The N and C termini of NS1 are denoted as N and C, respectively. β-Strands are numbered in the ED. The simulated annealing omit difference map contoured at ∼3σ for residues 71 to 84 in the H6N6-NS1 structure is shown as a gray mesh. Inset I shows a type I β-turn in the LR. Inset II shows intramolecular interactions between the RBD (orange) and the 170-loop of the ED (cyan). Residues involved in hydrogen bonding are shown as sticks, and hydrogen bonds are shown as dashed gray lines. (B) The NS1 dimer, with one subunit in cyan and the other in bright orange, is mediated by the RBD-RBD dimeric interactions, with the EDs positioned above the RBDs at an angle conferring the dimer a Y shape. An arrow indicates the area in which the LRs of each NS1 subunit in the dimer become intertwined. (C) Superposition of the H6N6-NS1 (cyan) and the VN/04 H5N1-NS1 (green) (PDB ID 3F5T) structures showing significant differences in their ED orientations with respect to RBD. Disordered residues (75 to 79) in the LR of VN/04 H5N1-NS1 (PDB ID 3F5T) structure are shown as a dotted green line.