ABSTRACT
The emergence of avian H7N9 viruses in humans in China has renewed concerns about influenza pandemics emerging from Asia. Vaccines are still the best countermeasure against emerging influenza virus infections, but the process from the identification of vaccine seed strains to the distribution of the final product can take several months. In the case of the 2009 H1N1 pandemic, a vaccine was not available before the first pandemic wave hit and therefore came too late to reduce influenza morbidity. H7 vaccines based on divergent isolates of the Eurasian and North American lineages have been tested in clinical trials, and seed strains and reagents are already available and can potentially be used initially to curtail influenza-induced disease until a more appropriately matched H7N9 vaccine is ready. In a challenge experiment in the mouse model, we assessed the efficacy of both inactivated virus and recombinant hemagglutinin vaccines made from seed strains that are divergent from H7N9 from each of the two major H7 lineages. Furthermore, we analyzed the cross-reactive responses of sera from human subjects vaccinated with heterologous North American and Eurasian lineage H7 vaccines to H7N9. Vaccinations with inactivated virus and recombinant hemagglutinin protein preparations from both lineages raised hemagglutination-inhibiting antibodies against H7N9 viruses and protected mice from stringent viral challenges. Similar cross-reactivity was observed in sera of human subjects from a clinical trial with a divergent H7 vaccine. Existing H7 vaccine candidates based on divergent strains could be used as a first line of defense against an H7N9 pandemic. In addition, this also suggests that H7N9 vaccines that are currently under development might be stockpiled and used for divergent avian H7 strains that emerge in the future.
IMPORTANCE Sporadic human infections with H7N9 viruses started being reported in China in the early spring of 2013. Despite a significant drop in the number of infections during the summer months of 2013, an increased number of cases has already been reported for the 2013-2014 winter season. The high case fatality rate, the ability to bind to receptors in the human upper respiratory tract in combination with several family clusters, and the emergence of neuraminidase inhibitor-resistant variants that show no loss of pathogenicity and the ability to transmit in animal models have raised concerns about a potential pandemic and have spurred efforts to produce vaccine candidates. Here we show that antigen preparations from divergent H7 strains are able to induce protective immunity against H7N9 infection.
INTRODUCTION
In the early spring of 2013, China reported the first human cases of avian H7N9 infections (1, 2) causing morbidity and high case fatality rates (3). Due to the emergence of resistant mutants, treatment with antivirals proved to be ineffective in a number of cases (4). Importantly, the mutations that confer resistance do not impact viral pathogenicity in the mouse model or the ability of the virus to transmit in the guinea pig model (5). Although no sustained human-to-human transmission has been detected so far (6), the ability of the hemagglutinin (HA) of this novel strain to bind weakly to alpha-2,6-linked sialic acid (7–11) suggests that H7N9 viruses could potentially become transmissible among humans. Subtype H7 viruses do not currently circulate in humans, and thus, any H7 virus that acquires the ability to spread from human to human could, if introduced into a naive population, cause a pandemic. For this reason, previous sporadic human infections with past H7 virus strains spurred the preclinical and early clinical development of prepandemic candidate vaccines (ClinicalTrials.gov registration numbers NCT00853255 and NCT00546585) (12, 13). These seed strains include H7 viruses of North American and Eurasian lineages (Fig. 1), with H7N9 being a member of the latter phylogenetic lineage. As it can take months to develop and evaluate novel matched vaccines based on H7N9, these preexisting vaccines, which might share conserved antigenic sites with recent H7N9 strains, could be used as a first line of defense against H7N9 viruses if these become pandemic before a matched vaccine is available.
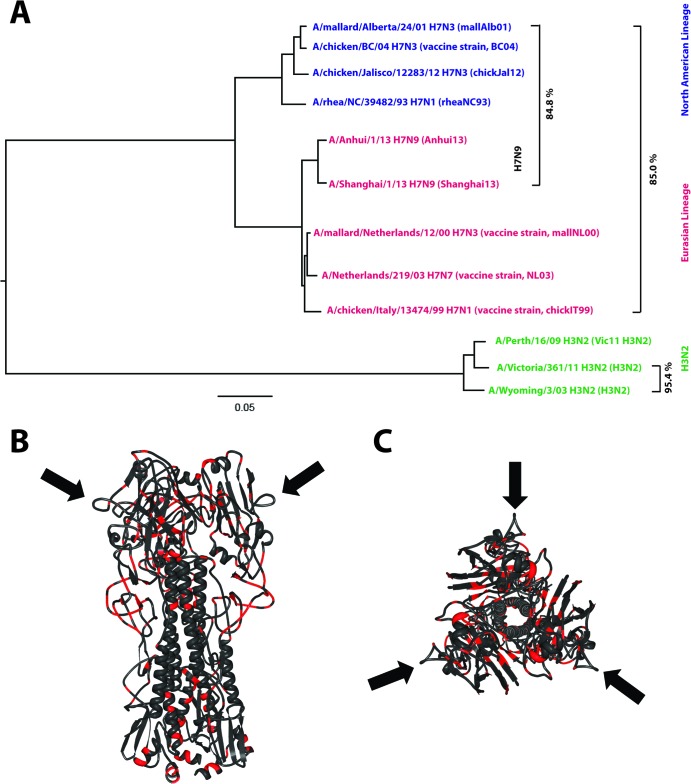
FIG 1.
Phylogenetic and antigenic relationship among H3 and H7 strains. (A) Phylogenetic tree based on HA sequences of North American (blue) and Eurasian (red) lineage H7 strains used in this study or in human clinical trials. H7N9 prototype strains are indicated. Drift in closely related H3N2 strains (95.4% amino acid identity between 2003 and 2009 isolates) mediates escape from HI-active antibodies quickly. HI cross-reactivity between distantly related H7 strains (e.g., mallAlb01 and Shanghai13 share only 84.8% amino acid identity) is probably mediated by conserved antigenic sites. The tree was built by using ClustalW and was visualized by using FigTree software. (B and C) Front view (B) and top view (C) of the HA trimer of A/Shanghai/2/13 (PDB accession number 4N5J [11]). Regions conserved among the vaccine strains tested in this study (Eurasian and North American lineages) are shown in dark gray, while nonconserved regions are shown in red. The completely conserved antigenic site A is indicated by black arrows.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK) cells (ATCC CCL-34) and human embryonic kidney 293T cells (ATCC CRL-11268) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) and minimal essential medium (Gibco), respectively. Both medium preparations were supplemented with 100 units/ml of 100 μg/ml penicillin and streptomycin (Pen/Strep; Gibco) and 10% fetal bovine serum (FBS; HyClone). The following wild-type and recombinant viruses were used in this study: A/rhea/North Carolina/39482/93 (H7N1) (rheaNC93); a 6:2 reassortant containing HA and NA of A/Shanghai/1/13 (H7N9) and the other genes from A/PR/8/34 (Shanghai13); a 6:2 reassortant containing HA and NA of A/Anhui/1/13 (H7N9) and other genes from A/PR/8/34 (Anhui13); a 7:1 reassortant containing HA of A/chicken/Jalisco/12283/12 (H7N3) lacking the polybasic cleavage site (low pathogenicity) and other genes from A/PR/8/34 (chickJal12); a 7:1 reassortant containing a chimeric HA with the globular head domain of A/mallard/Alberta/24/01 (H7N3) HA and the stalk domain from A/Perth/16/09 (H3N2) HA and other genes from A/PR/8/34 (mallAlb01); and a 7:1 reassortant containing HA of A/mallard/Netherlands/12/00 (H7N3) in combination with other genes from A/PR/8/34 (mallNL00), A/Victoria/361/11 (H3N2), and A/glaucous-winged gull/Southeastern Alaska/10JR01856R0/2010 (H11N9). Recombinant viruses were rescued as described previously (14); all viruses were grown in 10-day-old embryonated chicken eggs.
Vaccine preparation.
Viruses were grown in 10-day-old specific-pathogen-free embryonated eggs (Charles River), allantoic fluid was harvested and cleared by low-speed centrifugation (relative centrifugal force [RCF] of 2,000 for 10 min), and the virus was pelleted through a 30% sucrose cushion by ultracentrifugation (25,000 rpm for 2 h using a Beckman SW28 rotor). Virus pellets were resuspended in phosphate-buffered saline (PBS), inactivated with 0.03% formalin for 7 days at 4°C, and then dialyzed against PBS.
Recombinant proteins were produced in High Five insect cells by using the baculovirus expression system. All proteins used for vaccination contained a T4 trimerization domain to ensure correct folding and trimer formation of the proteins and a hexahistidine tag for purification (15, 16). The protein concentration of inactivated virus preparations and recombinant proteins was measured by using Bradford reagent (Bio-Rad).
Vaccination and challenge.
Animal protocols were reviewed and approved by the Mount Sinai Institutional Animal Care and Use Committee. Six- to eight-week-old BALB/c mice were vaccinated once or twice intramuscularly in the right hind leg with inactivated virus formulated in PBS (2 μg total protein per mouse), recombinant proteins [10 μg protein plus 10 μg poly(I·C) in PBS], or a commercially available trivalent inactivated vaccine (TIV) (Fluarix, 2010-2011 formulation, with 2 μg of HA from each strain for a total of 6 μg of HA, diluted in PBS). All vaccines were injected by using a 500-μl insulin syringe (BD). Another set of animals was intranasally infected with either 105 PFU of H3N2, H11N9, mallAlb01, or rheaNC93 or 0.1 50% lethal dose (LD50) of Shanghai13 in volumes of 50 μl. At 4 weeks postvaccination, animals were anesthetized, bled (submandibular), and challenged with 100 mouse LD50s (mLD50s) (1 mLD50 is equivalent to 530 PFU) of Shanghai13 virus. Weight was monitored daily for a period of 14 days; mice that lost 25% or more of their initial body weight were scored dead and euthanized. On days 3 and 6 postinfection, a subset of mice (n = 2 to 3 per group) was sacrificed, lungs were homogenized by using a FastPrep-24 instrument (MP Biomedicals), and lung titers were measured by a plaque assay on MDCK cells.
Human sera.
Human sera were obtained from a recent clinical trial with H7 vaccines at the University of Rochester (ClinicalTrials.gov registration number NCT01534468). In this trial, subjects received two doses of live attenuated A/Netherlands/219/2003 × A/Ann Arbor/6/60 (H7N7) cold-adapted vaccine (Medimmune) at a dose of 107.5 50% tissue culture infective doses (TCID50) administered by nasal spray 28 days apart. Approximately 18 months later, the subjects received a single dose of 45 μg of unadjuvanted subvirion H7N7 vaccine expressing HA of mallNL00 (Sanofi-Pasteur) intramuscularly (13). The tested sera were obtained prior to the inactivated booster dose and 14 days after the dose.
Enzyme-linked immunosorbent assay.
Enzyme-linked immunosorbent assay (ELISA) plates (Immunolon 4 HBX) were coated for 16 h at 4°C with purified protein (2 μg/ml) diluted in carbonate-bicarbonate coating buffer (pH 9.4). Plates were then blocked for 60 min at room temperature (RT) with PBS containing 0.1% Tween 20 (TPBS) and 3% nonfat dry milk powder (MTPBS). Serum was prediluted 1:50 and then serially diluted in 1:3 steps in MTPBS and incubated on the plates for 60 min at RT. Plates were then washed three times with 100 μl of TPBS per well and incubated for 60 min at RT with an anti-mouse IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz) diluted in MTPBS. Plates were then washed again with TPBS and developed by using the substrate o-phenylenediamine dihydrochloride (OPD) (SigmaFast; Sigma). Reactions were stopped by using 3 M HCl, and plates were read at an optical density of 490 nm. To prevent biased results when comparing sera from animals vaccinated with inactivated vaccines and recombinant proteins, we used recombinant Shanghai13 HA for ELISAs, which contained a different trimerization domain (GCN4pII) and tag (Strep-tag II) than those used for vaccination.
Hemagglutination inhibition assay.
Hemagglutination inhibition (HI) assays were conducted essentially as described previously (17, 18). HI titers for Shanghai13 and Anhui13 were determined with 0.5% turkey red blood cells, and HI titers for malNL00, rheaNC93, mallAlb01, and chickJal12 were determined by using 0.5% chicken red blood cells. HI titers are the reciprocal of the highest 2-fold dilution of the serum able to inhibit hemagglutination.
Enzyme-linked lectin assay allowing measurement of neuraminidase-inhibitory activity.
The neuraminidase activity of H7N9 was initially assessed by using a fetuin assay. Briefly, egg-grown virus was serially diluted with PBS-bovine serum albumin (BSA) in a final volume of 100 μl in a 96-well Maxisorp ELISA plate, and the assay was completed as described below. The dilution corresponding to the 50% effective dose (EC50) was determined by generating a nonlinear curve using Graphpad Prism 4. The EC50 dilution of virus (1:200) was then used to determine the amount of neuraminidase inhibition (NI) activity (described below). For the enzyme-linked lectin assay (ELLA), 96-well Maxisorp ELISA plates (Thermo-Scientific, Inc.) were coated with 150 μl (50 μg/ml) of fetuin (Sigma-Aldrich, Inc.) diluted in 0.1 M carbonate-bicarbonate buffer (pH 9.6) overnight at 4°C. The next day, plates were washed three times with 1× PBS and then blocked with 200 μl of 5% BSA–1× PBS for 1 h at RT. While plates were being blocked, 50 μl of diluted virus was coincubated with 50 μl of inactivated sera (serially diluted 2-fold) at RT for 1 h. After blocking, plates were washed three times with 0.5% Tween–1× PBS (TPBS), and 100 μl of the serum-virus mixture was then transferred to the fetuin-coated plates and incubated at 37°C for 2 h. The serum-virus mixture was then discarded, and plates were washed six times with 0.5% TPBS. One hundred microliters of peroxidase-conjugated peanut agglutinin (PNA; Sigma-Aldrich, Inc.) at 5 μg/ml was then added to the plates and incubated at RT for 2 h in the dark. Plates were again washed six times with 0.5% TPBS, and 100 μl of a peroxidase substrate, trimethylbenzidine (TMB), was added to each well of the plate. The reaction was stopped by using 50 μl of 3 M phosphoric acid, and the absorbance was read at 450 nm. The serum dilution corresponding to the 50% inhibitory concentration (IC50) was determined by generating a nonlinear curve using GraphPad Prism 6.
Statistical analysis, phylogenetic analysis, and visualization of molecular structures.
For statistical analysis of lung titers, a parametric unpaired t test was performed by using Prism 6 (GraphPad). Sequence alignments and trees were generated by using ClustalW, and trees were visualized with FigTree version 1.4.0. The molecular structures were visualized and modeled by using the Research Collaboratory for Structural Bioinformatics Protein Workshop.
RESULTS
Divergent H7 vaccines protect mice from H7N9 challenge.
We evaluated whether vaccines based on phylogenetically different Eurasian or North American lineage H7 strains (Fig. 1) induce cross-protective antibodies against H7N9 in the mouse model. Previous bioinformatics analyses predicted that the H7N9 strain might exhibit low immunogenicity and could act as a “stealth” virus due to its low content of T-cell epitopes (19). Therefore, we also evaluated the immunogenicity and protective efficacy of an inactivated H7N9 virus preparation in mice. Some degree of cross-reactivity between the distant North American and Eurasian lineage H7 strains has been reported previously and might be explained by shared, conserved antigenic sites (e.g., antigenic site A is 100% conserved between all H7 strains shown in Fig. 1) (20, 21).
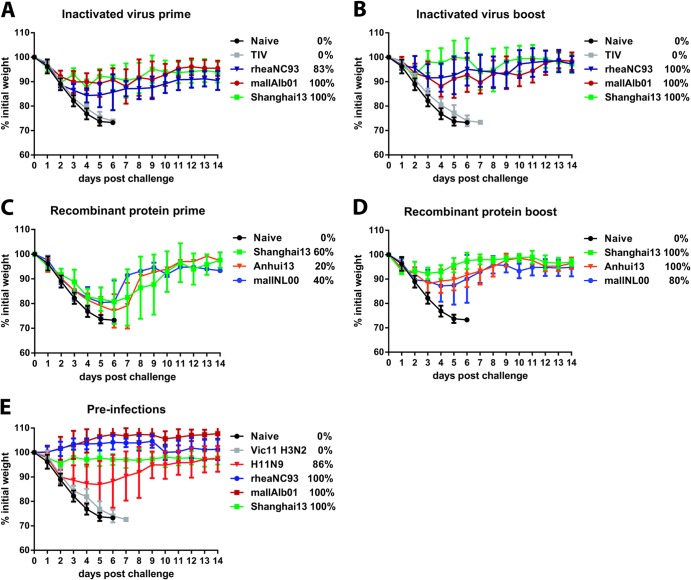
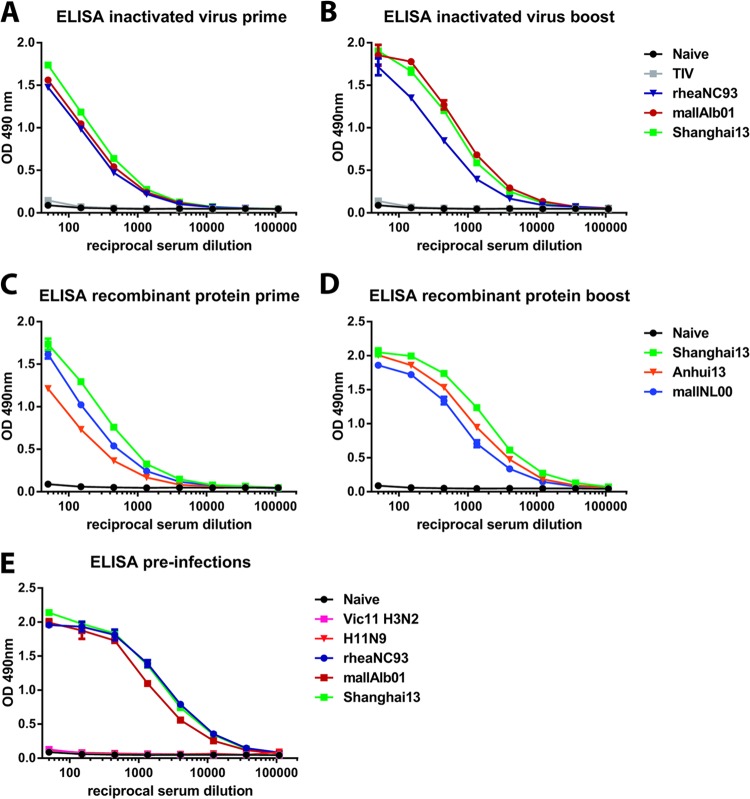
Animals vaccinated once with 2 μg (total protein) of inactivated virus preparations of North American H7 strains A/rhea/North Carolina/39482/93 (rheaNC93) and A/mallard/Alberta/24/01 (mallAlb01) lost approximately 15% of their body weight upon stringent challenge with 100 50% lethal doses (LD50) of A/Shanghai/1/13 (H7N9) (Shanghai13) virus and were completely (mallAlb01) or 83% (rheaNC93) protected from mortality, despite having very low to nondetectable HI titers against the challenge virus (Table 1 and Fig. 2A). Vaccination with inactivated Shanghai13 virus induced higher HI and ELISA titers against the challenge strain and against A/Anhui/1/13 (H7N9) (Anhui13) but did not significantly improve protection over levels seen with the heterologous vaccine preparations (Table 1 and Fig. 2A). A second vaccine dose boosted HI titers against Shanghai13 and Anhui13 viruses, boosted the reactivity to Shanghai13 HA in ELISAs, and alleviated weight loss compared to animals that received the vaccine just once (Fig. 2B and 3A and B). The group vaccinated with homologous Shanghai13 virus lost less weight on days 3 to 6, but differences were not statistically significant. Survival was 100% in all H7 prime-boost groups (Fig. 2B).
TABLE 1.
Hemagglutination inhibition titers of mice vaccinated against two prototype H7N9 strains
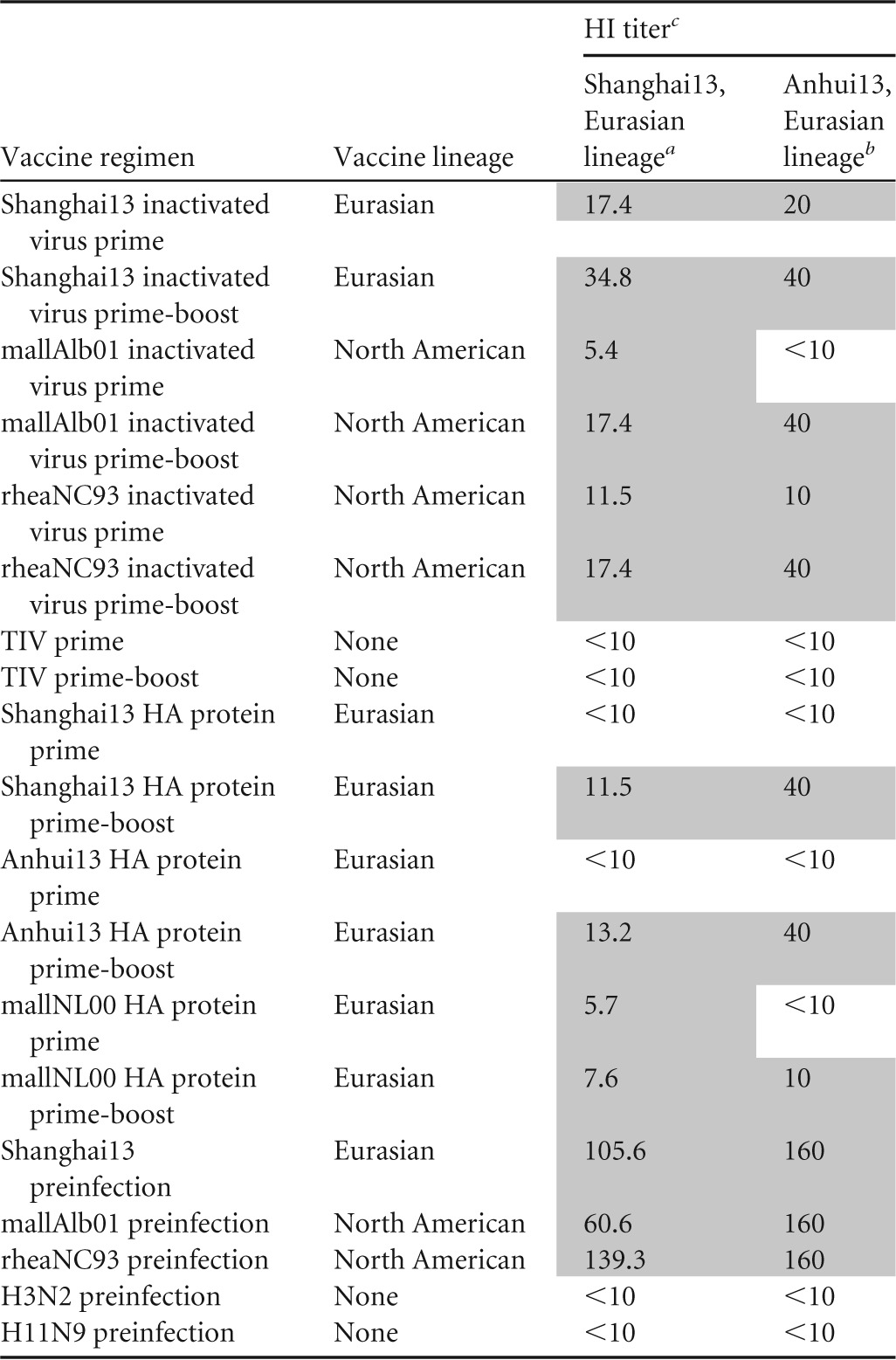
Sera were tested individually for reactivity against Shanghai13, negative sera (titer of <10) were assigned a value of 5, and geometric mean titers were calculated.
Pooled sera (n = 5 to 10) were tested.
<10 indicates that no HI activity was detected.
FIG 2.
Exposure to divergent H7 antigens protects mice against H7N9 challenge. (A) Weight loss of animals vaccinated once with inactivated H7 viruses derived from the North American (mallAlb01 and rheaNC93) or Eurasian (Shanghai13, homologous) lineage (n = 5 to 6 for vaccine groups; n = 17 for naive animals). (B) Weight loss of animals vaccinated twice with inactivated H7 viruses derived from the North American (mallAlb01 and rheaNC93) or Eurasian (Shanghai13, homologous) lineage (n = 5 for vaccine groups; n = 17 for naive animals, which are the same as those shown in panel A). (C) Weight loss of animals vaccinated once with recombinant H7 HA protein (mallNL00, Shanghai13, and Anhui13) (n = 5 for vaccine groups; n = 17 for naive animals, which are identical to the animals shown in panel A). (D) Weight loss of animals vaccinated twice with recombinant H7 HA protein (mallNL00, Shanghai13, and Anhui13) (n = 5 for vaccine groups; n = 17 for naive animals, which are the same as those shown in panel A). (E) Weight loss of animals preinfected with divergent H7 subtype viruses of the North American (rheaNC93 and mallAlb01) or Eurasian (Shanghai13) lineage, an H11N9 virus, or an H3N2 virus (n = 5 to 7 for preinfection groups; n = 17 for naive animals, which are identical to the animals shown in panel A). Survival rates are indicated in the keys as percentages.
FIG 3.
ELISA reactivity of vaccinated or infected animals to recombinant H7 hemagglutinin. Shown is ELISA reactivity of sera from animals vaccinated once (A) or twice (B) with inactivated vaccines or vaccinated once (C) or twice (D) with recombinant HA antigen or of sera from animals preinfected with divergent influenza viruses (E). Recombinant H7 HA derived from Shanghai13 was used as the substrate. To rule out a biased readout, the substrate was expressed with a different trimerization domain and purification tag (GCN4pII trimerization domain and Strep-tag II) than the recombinant HA used for vaccination (T4 fibritin trimerization domain and hexahistidine tag). OD, optical density.
We also determined lung titers on days 3 and 6 after challenging the mice that had been vaccinated once or twice with an inactivated virus vaccine and detected no or low viral titers compared to those of naive animals, which exhibited titers of about 106 PFU/ml (Fig. 4C and D). No statistically significant difference was observed in lung virus titers measured in mice vaccinated with inactivated rheaNC93 or mallAlb01 virus compared to those in mice vaccinated with Shanghai13 virus in either of the two vaccination regimens or two time points. The only exception was detected for the prime-boost regimen, where rheaNC93-vaccinated animals showed significantly lower titers on day 6 than did Shanghai13-vaccinated animals (P = 0.0025). Due to the small number of animals per condition (n = 2) in the prime-only mallAlb01 day 3 and the prime-only rheaNC93 day 6 groups, we could not perform a statistical analysis for significance of the differences between these groups and the prime-only Shanghai13 animals.
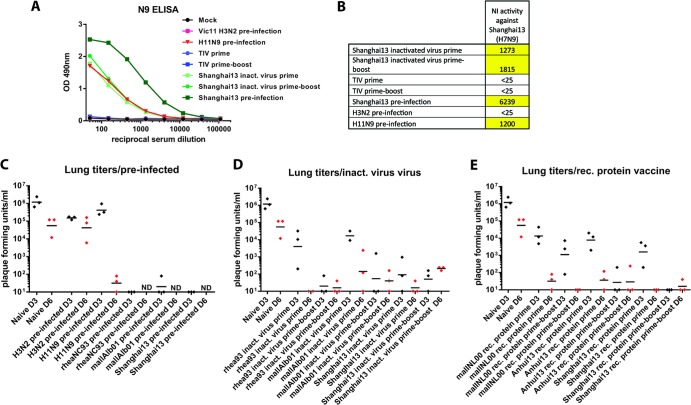
FIG 4.
Neuraminidase inhibition activity and viral lung titers in mice. (A) ELISA reactivity of selected groups to N9 NA. (B) N9 NI titers of groups shown in panel C. NI was measured by using A/Shanghai/1/13 virus; values are expressed as the reciprocal serum dilution that was able to inhibit NA activity by 50%. (C to E) Lung titers of mice vaccinated with different vaccination regimens on day 3 (black) and day 6 (red) postinfection. Samples with no detectible virus titer were scored as 10. The naive day 3 and day 6 groups were included in all three panels to facilitate comparison with the vaccine groups. ND, not determined.
Mice vaccinated once or twice with a commercial trivalent inactivated vaccine (TIV) containing H1, H3, and influenza B virus components were not protected from infection, suggesting that regular influenza virus vaccines lack efficacy against the novel Chinese H7N9 virus. However, animals that received the TIV twice showed slightly delayed weight loss kinetics and did not show signs of enhanced pathogenicity (Fig. 2A and B). It is of note that the amount of internal proteins that can induce T-cell-mediated cross-protection is probably smaller in the commercial TIV than in the inactivated whole-virus vaccine preparations used in this study. However, TIV-vaccinated mice received higher doses of HA (2 μg/mouse) than animals that received whole-virus inactivated vaccines (2 μg of total protein).
Recombinant HA (rHA)-based influenza virus vaccines were recently licensed for use in humans (22) and can potentially enhance pandemic preparedness by shortening the time between vaccine strain selection and production. It is therefore important to evaluate if such a vaccine approach can be used for H7N9. Protein for this evaluation was produced in an insect cell expression system capable of performing posttranslational modifications, and the structural integrity of the recombinant proteins was tested by using monoclonal antibody CR9114 (16, 23), which binds to a conformational epitope in the stalk domain of HA (Fig. 5B). Animals vaccinated once with an adjuvanted dose of homologous Shanghai13 or Anhui13 or heterologous A/mallard/Netherlands/12/00 (mallNL00) (Eurasian H7 lineage) rHA protein (Fig. 5) did not mount HI titers against Shanghai13 and showed only partial protection against viral challenge, although all groups seroconverted, as measured by ELISA (Table 1 and Fig. 2C and 3C). Animals vaccinated with a second dose of recombinant vaccine had a boost in HI and ELISA titers and were fully (Shanghai13 and Anhui13) or partially (mallNL00) (80%) protected from mortality and morbidity (Table 1 and Fig. 2D). Similar to the results from the inactivated vaccines, we did not detect significant differences between lung titers of animals vaccinated with heterologous protein and those of animals that received homologous Shanghai13 protein. The mallNL00 protein, however, seemed to provide less protection than the Shanghai13 and Anhui13 proteins in the prime-boost regimen on day 3 (Fig. 4E).
FIG 5.
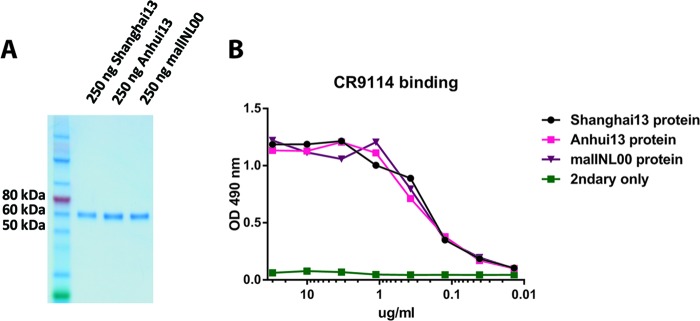
Characterization of recombinant H7 hemagglutinin proteins. (A) Coomassie staining of an SDS-PAGE gel with the three different H7 HAs used in the vaccination study. (B) ELISA with stalk-reactive antibody CR9114. This monoclonal antibody binds to a conformational epitope in the HA stalk and was used to assess the structural integrity of the proteins used.
Humans mount cross-reactive HI responses to H7N9 virus upon vaccination with a divergent H7 vaccine.
To validate the results from the mouse model, we obtained six available sera from a human clinical trial with H7 vaccines. Samples were tested for HI reactivity to Shanghai13 and Anhui13 H7N9 viruses as well as a panel of other H7 viruses of both lineages. Similar to observations in the mouse model, individuals who seroconverted to the vaccine strain (Table 2) also showed high cross-reactivity to H7N9 viruses. Individuals who showed no titer against the vaccine strain (patients 42 and 43) (Table 2) also showed no reactivity to H7N9, whereas individuals who showed high titers against the vaccine strain (patients 44, 45, and 46) showed similarly high titers against H7N9 viruses and divergent viruses of the North American lineage. This suggests that the observed cross-reactivity is not mouse specific and that the human immune system targets H7 epitopes similarly to the mouse immune system. In contrast, recent data from the ferret model showed a very narrow HI response to specific H7 strains, a phenomenon which has also been observed for H1N1 strains (24, 25).
TABLE 2.
HI titers of six vaccinees against heterologous H7 viruses, including two prototype H7N9 strains and the homologous vaccine virus strain
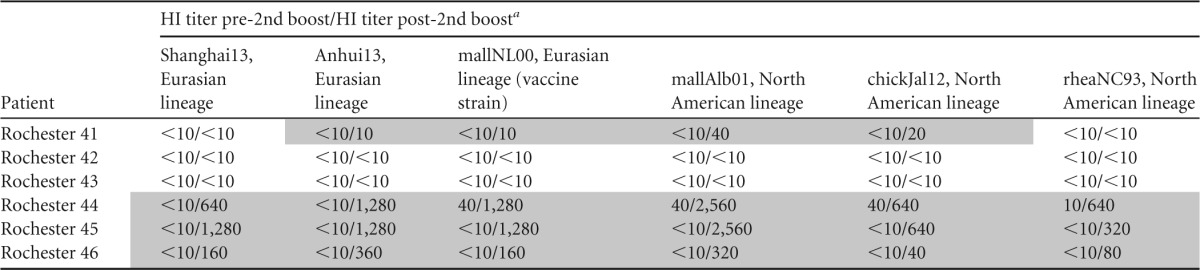
Vaccinees received a cold-adapted, attenuated vaccine expressing HA of A/Netherlands/219/03 twice (28 days apart) and were then boosted approximately 18 months later with an A/mallard/NL/12/00 (mallNL00) inactivated vaccine containing 45 μg of HA. The tested sera were obtained prior to the inactivated booster dose (pre-2nd dose, the first value presented in each cell) and 14 days after the dose (post-2nd dose, the second value presented in each cell).
Preexposure to divergent H7 and H11N9 but not H3N2 viruses protects mice from H7N9 challenge.
As H3 and H7 HAs both belong to the same phylogenetic group and H3N2 viruses circulate in the human population, we investigated whether preexisting immunity induced by exposure to the H3 subtype had a positive or negative impact on the pathogenicity of the H7N9 virus following challenge. We preinfected animals with an H3N2 strain and several H7 strains, including Shanghai13 for comparison. A group of mice preinfected with an H11N9 strain (seroconverted to N9 NA) (Fig. 4A and B) of North American avian origin was selected to determine if an immune response against NA would be sufficient to protect mice from challenge (94% amino acid similarity to N9 NA of Shanghai13). Animals that experienced H7 preinfection mounted high HI titers against Shanghai13 and Anhui13 and high ELISA reactivity to Shanghai13 protein and were completely protected from weight loss and mortality (Table 1 and Fig. 2E and 4E). H11N9-preinfected animals lost approximately 15% of their initial body weight and were protected from mortality (86% survived) while showing no reactivity to H7 HA (Fig. 4E). H3N2-preinfected animals (animals seroconverted to H3 HA) (data not shown) showed no enhanced protection compared to naive mice in terms of weight loss or mortality but had reduced lung titers (approximately 105 PFU, compared to 106 PFU in naive mice) (Fig. 4C). Thus, preexisting immunity does not induce enhanced pathogenicity in this model, which correlates with TIV data (Fig. 2A and B). The reduced lung titers in H3N2-preinfected mice are most likely caused by cross-reactive T-cell responses to conserved internal influenza virus proteins. With the exception of one animal from the mallAlb01 group, infectious challenge viruses could not be detected in lungs of any of the H7-preinfected mice (Fig. 4C); H11N9-preinfected animals showed no reduction in lung virus titers on day 3, but only low titers of residual virus were found on day 6 postchallenge (Fig. 4C).
Vaccine-induced anti-N9 antibodies efficiently inhibit H7N9 neuraminidase.
Since H11N9-preinfected animals were protected from challenge, we also assessed the levels of humoral immunity against N9 NA induced by preinfection and vaccination. Preinfection of animals with Shanghai13 virus induced high titers of anti-N9 antibodies, as measured by ELISA, and also showed a high neuraminidase inhibition (NI) titer of 1:6,239 (Fig. 4A and B). Inactivated Shanghai13 virus administered once or twice, as well as preinfection with H11N9, also induced anti-N9 reactivity and good NI activity (Fig. 4A and B). It is of note that the anti-N9 ELISA titer, as well as the NI titer, was not boosted by a second vaccination.
DISCUSSION
Subtype H7 influenza viruses have sporadically caused disease in humans (26–30). In general, these infections are associated with mild symptoms, including conjunctivitis, but can also lead to more serious outcomes, including death (27). H7 vaccine candidates based on North American and Eurasian lineage virus strains (Fig. 1A) have been developed and tested preclinically and clinically. Recently, severe human cases of avian H7N9 infections with high case fatality rates in China raised concerns that this virus could become pandemic. Sequence analysis and biochemical studies revealed the presence of mutations in the HA gene that enable the virus to bind slightly to alpha-2,6-linked sialic acid (7, 11), a prerequisite for efficient spread among human individuals. Furthermore, several isolates showed resistance to antiviral drugs due to the presence of the R294K mutation in the viral NA (4), a mutation that was not associated with an attenuated viral phenotype in the H7N9 background (5, 8).The development of a prepandemic vaccine against this novel H7N9 strain is therefore of the highest priority.
We tested a matched vaccine based on an inactivated A/Shanghai/1/13 virus reassortant, which proved to be as immunogenic as H1N1-inactivated vaccines in mice (31), suggesting that inactivated H7N9 vaccines could also be immunogenic in humans. Inactivated mismatched H7 vaccines from the two distinct H7 lineages also conferred good protection. Recombinant HA vaccines have recently been licensed for use in humans and can be produced more rapidly than classical inactivated virus vaccines (22). We tested several recombinant H7 HAs for their ability to protect animals from challenge and found that in most cases, a single vaccination was insufficient to protect from viral infection, even when the antigen was matched. However, robust protection was observed after animals received a recombinant HA boost. It has been speculated that preexisting immunity induced by influenza virus vaccination or infection might increase the pathogenicity of H7N9 in the elderly (32). We found no evidence of increased morbidity or mortality due to preexisting immunity to human seasonal strains in the mouse model. Although TIV-vaccinated or H3N2-infected mice were not protected from mortality, they showed slightly delayed weight loss kinetics and, in some cases, slightly reduced lung titers compared to naive mice, an effect that is most likely caused by cross-reactive cytotoxic T cells (33). Antibodies against N9 NA, which are usually nonneutralizing, provided notable levels of NI activity and were able to partially protect animals from challenge with H7N9 virus. This also indicates that an ideal vaccine should include an N9 component to maximize protection.
As happened during the 2009 H1N1 pandemic response, the development of a vaccine based on a novel strain usually takes several months and might come too late if the virus acquires sustained human-to-human transmission. Our results suggest that there is high cross-reactivity between divergent H7 strains of both lineages, which enables robust protection from H7N9 challenge in mice, even after only one vaccine dose and with low serum HI titers. This is an important finding since H7 vaccines in humans proved to be of low immunogenicity when HI titers were used as a readout and also suggests that other types of antibodies, such as non-HI-active/nonneutralizing antibodies or neutralizing antistalk antibodies, might contribute to protection (34). There are multiple H7 vaccine strains, such as A/Netherlands/219/03 (H7N7), A/chicken/British Columbia/2004 (H7N3), A/mallard/Netherlands/24/00 (H7N3), A/chicken/Italy/13474/99 (H7N1), and others, that have already been tested in humans (ClinicalTrials.gov registration numbers NCT00853255 and NCT00546585) (12, 13, 35). Sera from individuals from one of these trials were tested for HI reactivity to heterologous strains, including two representative H7N9 viruses, and strong cross-reactivity in seroconverted individuals was confirmed.
Our data therefore suggest that existing H7 vaccine candidates based on divergent strains could be employed in case of an H7N9 pandemic. These results and recent reports from other groups show that there is substantial cross-reactivity between H7 strains in mice and humans (20, 36–39) but to a much lesser extent in ferrets (24). This cross-reactivity is most likely induced by antibodies against antigenic site A, which is highly conserved in avian H7 isolates. Various adjuvanted and nonadjuvanted H7N9 vaccines that are now in phase I (ClinicalTrials.gov registration numbers NCT01934127, NCT01995695, and NCT01928472) (40) and phase II (ClinicalTrials.gov registration number NCT01942265) clinical trials could therefore also be stockpiled and likely be used against divergent avian H7 strains that emerge in the human population in the future.
ACKNOWLEDGMENTS
We thank Ariana Hirsh and Chen Wang for excellent technical support and Amy and Edward Nisselle for editing of the manuscript.
F.K. was supported by an Erwin Schrödinger fellowship (J3232) from the Austrian Science Fund (FWF). This work was partially supported by a Centers for Excellence for Influenza Research and Surveillance (CEIRS) grant (grant HHSN26620070010C to A.G.-S., P.P., J.J.T., and J.R.), NIH program project grant 1P01AI097092-01A1 (to A.G.-S., P.P., and P.C.W.), PATH (A.G.-S. and P.P.), and NIH contracts HHSN272200900026C (J.J.T.) and HHSN272201000054C (A.G.-S. and R.A.A.).
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 2.CDC. 2013. Emergence of avian influenza A(H7N9) virus causing severe human illness—China, February-April 2013. MMWR Morb. Mortal. Wkly. Rep. 62:366–371 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6218a6.htm [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, Zhang H, Li M, Zeng L, Xu Z, Li Z, Luo H, Li Q, Feng Z, Cao B, Yang W, Wu JT, Wang Y, Leung GM. 2013. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 382:138–145. 10.1016/S0140-6736(13)61207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. 2013. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 381:2273–2279. 10.1016/S0140-6736(13)61125-3 [DOI] [PubMed] [Google Scholar]
- 5.Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solórzano A, García-Sastre A, Palese P, Bouvier NM. 2013. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat. Commun. 4:2854. 10.1038/ncomms3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ip DK, Liao Q, Wu P, Gao Z, Cao B, Feng L, Xu X, Jiang H, Li M, Bao J, Zheng J, Zhang Q, Chang Z, Li Y, Yu J, Liu F, Ni MY, Wu JT, Cowling BJ, Yang W, Leung GM, Yu H. 2013. Detection of mild to moderate influenza A/H7N9 infection by China's national sentinel surveillance system for influenza-like illness: case series. BMJ 346:f3693. 10.1136/bmj.f3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. 10.1038/nature12372 [DOI] [PubMed] [Google Scholar]
- 8.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, García-Sastre A, Fernandez-Sesma A. 2013. H7N9 influenza viruses interact preferentially with α2,3-linked sialic acids and bind weakly to α2,6-linked sialic acids. J. Gen. Virol. 94:2417–2423. 10.1099/vir.0.056184-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. 2013. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342:1230–1235. 10.1126/science.1243761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Höschler K, Saville M, Vogel FR, Barclay W, Donatelli I, Zambon M, Wood J, Haaheim LR. 2009. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine 27:1889–1897. 10.1016/j.vaccine.2009.01.116 [DOI] [PubMed] [Google Scholar]
- 13.Couch RB, Patel SM, Wade-Bowers CL, Niño D. 2012. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One 7:e49704. 10.1371/journal.pone.0049704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, Palese P. 2012. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J. Virol. 86:5774–5781. 10.1128/JVI.00137-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7:e43603. 10.1371/journal.pone.0043603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margine I, Palese P, Krammer F. 2013. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp. 2013:e51112. 10.3791/51112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Taaffe J, Parker C, Solórzano A, Cao H, García-Sastre A, Lu S. 2006. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. J. Virol. 80:11628–11637. 10.1128/JVI.01065-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, Xu H, Pascual V, Banchereau J, Garcia-Sastre A, Palucka AK, Ramilo O, Ueno H. 2013. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 5:176ra32. 10.1126/scitranslmed.3005191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Groot AS, Ardito M, Terry F, Levitz L, Ross TM, Moise L, Martin W. 2013. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum. Vaccin. Immunother. 9:950–956. 10.4161/hv.24939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff PH, Krammer F, Hai R, Seibert CW, Margine I, García-Sastre A, Palese P. 2013. Induction of cross-reactive antibodies to novel H7N9 influenza virus by recombinant Newcastle disease virus expressing a North American lineage H7 subtype hemagglutinin. J. Virol. 87:8235–8240. 10.1128/JVI.01085-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbas MA, Spackman E, Fouchier R, Smith D, Ahmed Z, Siddique N, Sarmento L, Naeem K, McKinley ET, Hameed A, Rehmani S, Swayne DE. 2011. H7 avian influenza virus vaccines protect chickens against challenge with antigenically diverse isolates. Vaccine 29:7424–7429. 10.1016/j.vaccine.2011.07.064 [DOI] [PubMed] [Google Scholar]
- 22.US Food and Drug Administration. 2013. FDA approves new seasonal influenza vaccine made using novel technology. US Food and Drug Administration, Washington, DC: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm335891.htm [Google Scholar]
- 23.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin Ö, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q, Chen Z, Cheng X, Xu L, Jin H. 2013. Evaluation of live attenuated H7N3 and H7N7 vaccine viruses for their receptor binding preferences, immunogenicity in ferrets and cross reactivity to the novel H7N9 virus. PLoS One 8:e76884. 10.1371/journal.pone.0076884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, Principi N, Plotkin JB, Ross TM, Ahmed R, Wilson PC, Hensley SE. 2013. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 210:1493–1500. 10.1084/jem.20130212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 10:2196–2199. 10.3201/eid1012.040961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361. 10.1073/pnas.0308352100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CDC. 2012. Notes from the field: highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers—Jalisco, Mexico, July 2012. MMWR Morb. Mortal. Wkly. Rep. 61:726–727 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6136a4.htm [PubMed] [Google Scholar]
- 29.Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg. Infect. Dis. 15:859–865. 10.3201/eid1506.090072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morens DM, Taubenberger JK, Fauci AS. 2013. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. mBio 4(4):e00445–13. 10.1128/mBio.00445-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villán E, Palese P, Basler CF, García-Sastre A. 2010. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 6:e1000745. 10.1371/journal.ppat.1000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skowronski D, Janjua N, Kwindt T, De Serres G. 2013. Virus-host interactions and the unusual age and sex distribution of human cases of influenza A(H7N9) in China, April 2013. Euro Surveill. 18(17):20465 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20465 [PubMed] [Google Scholar]
- 33.van de Sandt CE, Kreijtz JH, de Mutsert G, Geelhoed-Mieras MM, Hillaire ML, Vogelzang-van Trierum SE, Osterhaus AD, Fouchier RA, Rimmelzwaan GF. 2014. Human cytotoxic T lymphocytes directed to seasonal influenza A viruses cross-react with the newly emerging H7N9 virus. J. Virol. 88:1684–1693. 10.1128/JVI.02843-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krammer F, Cox RJ. 2013. The emergence of H7N9 viruses: a chance to redefine correlates of protection for influenza virus vaccines. Expert Rev. Vaccines 12:1369–1372. 10.1586/14760584.2013.850036 [DOI] [PubMed] [Google Scholar]
- 35.Cox RJ, Major D, Hauge S, Madhun AS, Brokstad KA, Kuhne M, Smith J, Vogel FR, Zambon M, Haaheim LR, Wood J. 2009. A cell-based H7N1 split influenza virion vaccine confers protection in mouse and ferret challenge models. Influenza Other Respir. Viruses 3:107–117. 10.1111/j.1750-2659.2009.00082.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith GE, Flyer DC, Raghunandan R, Liu Y, Wei Z, Wu Y, Kpamegan E, Courbron D, Fries LF, Glenn GM. 2013. Development of influenza H7N9 virus like particle (VLP) vaccine: homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA1/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine 31:4305–4313. 10.1016/j.vaccine.2013.07.043 [DOI] [PubMed] [Google Scholar]
- 37.Rudenko L, Isakova-Sivak I, Donina S. 2013. H7N3 live attenuated influenza vaccine has a potential to protect against new H7N9 avian influenza virus. Vaccine 31:4702–4705. 10.1016/j.vaccine.2013.08.040 [DOI] [PubMed] [Google Scholar]
- 38.Rudenko L, Isakova-Sivak I, Rekstin A. 2014. H7N9: can H7N3 live-attenuated influenza vaccine be used at the early stage of the pandemic? Expert Rev. Vaccines 13:1–4. 10.1586/14760584.2014.864564 [DOI] [PubMed] [Google Scholar]
- 39.Klausberger M, Wilde M, Palmberger D, Hai R, Albrecht RA, Margine I, Hirsh A, García-Sastre A, Grabherr R, Krammer F. 2014. One-shot vaccination with an insect cell-derived low-dose influenza A H7 virus-like particle preparation protects mice against H7N9 challenge. Vaccine 32:355–362. 10.1016/j.vaccine.2013.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fries LF, Smith GE, Glenn GM. 2013. A recombinant viruslike particle influenza A (H7N9) vaccine. N. Engl. J. Med. 369:2564–2566. 10.1056/NEJMc1313186 [DOI] [PubMed] [Google Scholar]