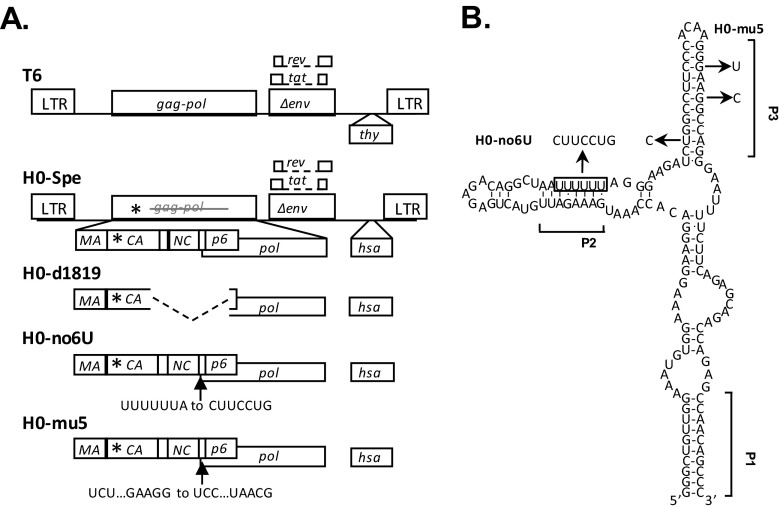
FIG 1.
General structures of HIV-1 constructs (A) and predicted RNA structure of the HIV-1 Gag-Pol ribosomal frameshift signal (B). Asterisks denote locations of the stop codon insertion in the CA-coding region. Dashed lines indicate deletions, and arrows indicate the locations of point mutations. The predicted secondary structure of NL4-3 ribosomal frameshift signal is modified from an earlier study (42), with square brackets indicating positions of proposed RNA helices P1, P2, and P3. The boxed sequence denotes the stretch of six U nucleotides that constitute the ribosome slippery sequence. The locations of point mutations and the names of the mutants are shown. Three nucleotide changes were introduced into the 6 U stretch to generate H0-no6U, whereas three nucleotide mutations were introduced into the P3 helix to generate H0-mu5.