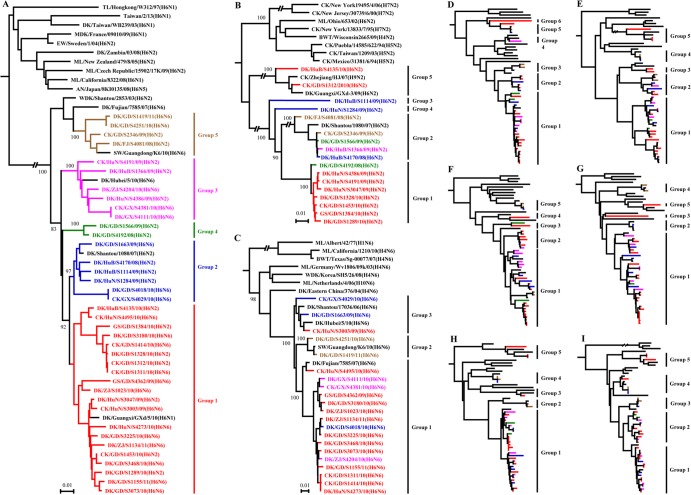
FIG 2.
Phylogenetic analyses of the H6 viruses isolated from 2008 to 2011 in China. The phylogenetic trees were generated with the PHYLIP program of the ClustalX software package (version 1.81). The nine trees were generated based on the following sequences: HA nucleotides (nt) 18 to 1718, N2 nt 20 to 1429, N6 nt 20 to 1432, PB2 nt 28 to 2307, PB1 nt 25 to 2298, PA nt 25 to 2175, NP nt 46 to 1542, M nt 26 to 1007, and NS nt 27 to 864. The phylogenetic trees of HA (A), N2 NA (B), and N6 NA (C) were rooted to A/Turkey/Canada/1963 (H6N2), A/Turkey/England/1969 (H3N2), and A/Duck/England/1/1956 (H11N6), respectively. The phylogenetic trees of PB2 (D), PB1 (E), PA (F), NP (G), M (H), and NS (I) were rooted to A/Equine/Prague/1/56 (H7N7). Sequences of viruses with names in black were downloaded from available databases; viruses with names in other colors were sequenced in this study. The colors of the virus names in the NA, PB2, PB1, PA, NP, M, and NS trees match with those used in the HA tree. Abbreviations: AN, avian; BWT, blue-winged teal; CK, chicken; DK, duck; EC, Eastern China; EW, Eurasian wigeon; FJ, Fujian; GD, Guangdong; GS, goose; GX, Guangxi; HK, Hongkong; HuB, Hubei; HuN, Hunan; MDK, Muscovy duck; ML, mallard; SW, swine; TL, teal; WDK, wild duck; ZJ, Zhejiang.