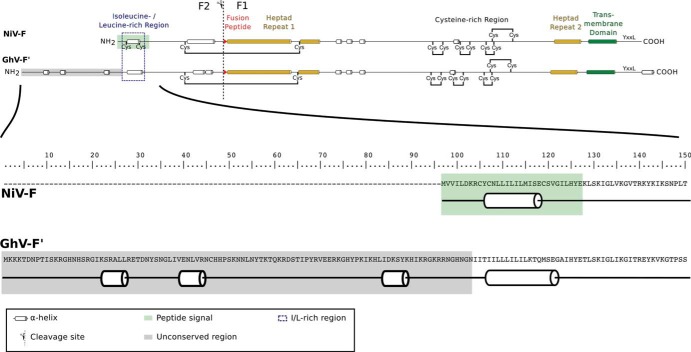
FIG 2.
Comparison of conserved features in NiV-F and GhV-F′. (Top) Comparison of conserved features across the full length of NiV-F and GhV-F′. The software PSIPRED was used to predict the secondary structure of the NiV-F and GhV-F′ protein sequences. A schematic of the two proteins highlights major features and conserved regions. The unconserved region of the N terminus in GhV-F′ consists of about 100 amino acids. (Bottom) Amino acid sequence of the full N-terminal region of GhV-F′. An extended version of Fig. 2A is shown, which includes the complete amino acid sequence of the unconserved N-terminal region of GhV-F′.