FIG 7.
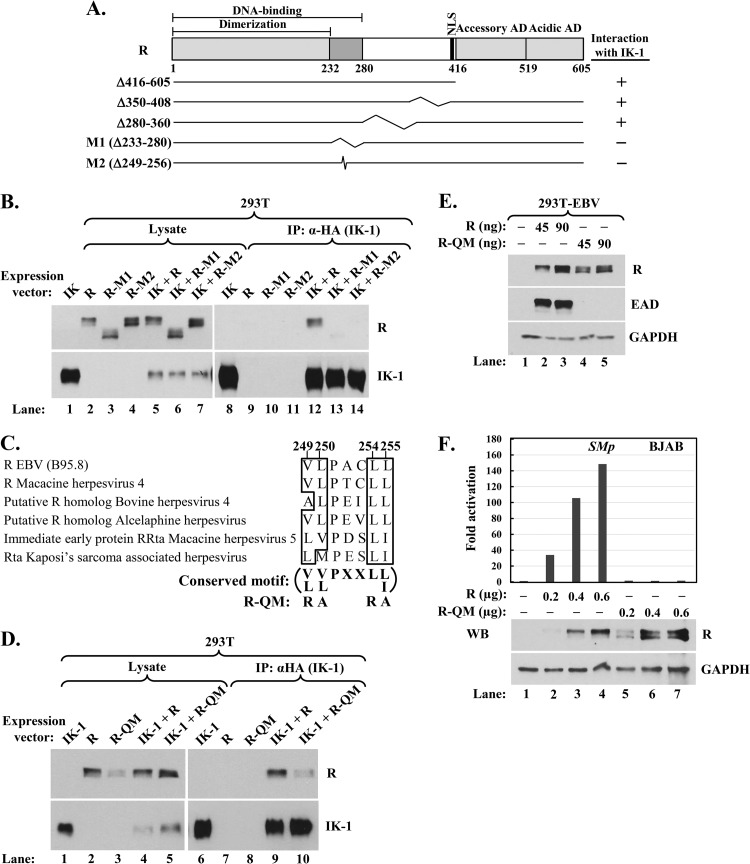
Conserved hydrophobic amino acid residues 249, 250, 254, and 255 of R are critical for its interaction with Ikaros. (A) Schematic showing R's DNA-binding, dimerization, nuclear localization (NLS), and accessory and acidic activation domains (AD). Numbers indicate amino acid residues. Deletion mutants analyzed in coimmunoprecipitation assays are shown; kinks denote internally deleted regions. (B) Immunoblot showing coimmunoprecipitation of R mutant variants with IK-1. 293T cells in 6-well plates were cotransfected as follows: lanes 1 and 8, 0.28 μg pcDNA3-HA-IK-1; lanes 2 and 9, 0.25 μg pcDNA3-R; lanes 3 and 10, 0.45 μg pcDNA3-R-M1; lanes 4 and 11, 0.30 μg pcDNA3-R-M2; lanes 5 and 12, 0.31 μg pcDNA3-HA-IK-1 plus 0.25 μg pcDNA3-R; lanes 6 and 13, 0.25 μg pcDNA3-HA-IK-1 plus 0.45 μg pcDNA3-R-M1; and lanes 7 and 14, 0.28 μg pcDNA3-HA-IK-1 plus 0.30 μg pcDNA3-R-M2; total DNA was brought up to 0.70 μg per well with pcDNA3.1 where needed. Whole-cell extracts were prepared 48 h later, and complexes were coimmunoprecipitated with anti-HA tag antibody. (C) Alignment of amino acid residues 248 to 256 of EBV R with similar residues from the R-like proteins of some other gamma herpesviruses. Conserved hydrophobic residues are emphasized by boxes. The substitution mutations present in quadruple mutant R-QM are shown. (D) Immunoblot showing reduced coimmunoprecipitation of mutant R-QM with IK-1. 293T cells in 6-well plates were cotransfected as follows: lanes 1 and 6, 0.20 μg pcDNA3-HA-IK-1; lanes 2 and 7, 0.20 μg pcDNA3-R; lanes 3 and 8, 0.20 μg pcDNA3-R-QM; lanes 4 and 9, 0.36 μg pcDNA3-HA-IK-1 plus 0.20 μg pcDNA3-R; and lanes 5 and 10, 0.36 μg pcDNA3-HA-IK-1 plus 0.20 μg pcDNA3-R-QM; total DNA was brought up to 0.56 μg per well with pcDNA3.1 where needed. Whole-cell extracts were prepared and processed as described in the legend for panel B. (E) Immunoblot showing failure of mutant R-QM to disrupt EBV latency. 293T-EBV cells in a 12-well plate were transfected with the indicated amounts of pcDNA3-R or pcDNA3-R-QM plus pcDNA3.1 to bring total DNA to 0.3 μg per well and were harvested 48 h later. (F) Luciferase reporter assays showing failure of mutant R-QM to activate the EBV SM (BMLF1) promoter. BJAB cells were coelectroporated with 1.7 μg pCpGL-SMp reporter plasmid, 0.4 μg pcDNA3-eGFP, and the indicated amounts of pcDNA3-R or pcDNA3-R-QM (plus vector pcDNA3.1 to bring total DNA to 2.7 μg per sample). Luciferase activities were determined 44 h later. Data were normalized internally to the amount of protein in each lysate and externally to basal activity observed in the absence of R. Immunoblot analysis was also performed to determine WT and mutant R protein levels. WB, Western blot.