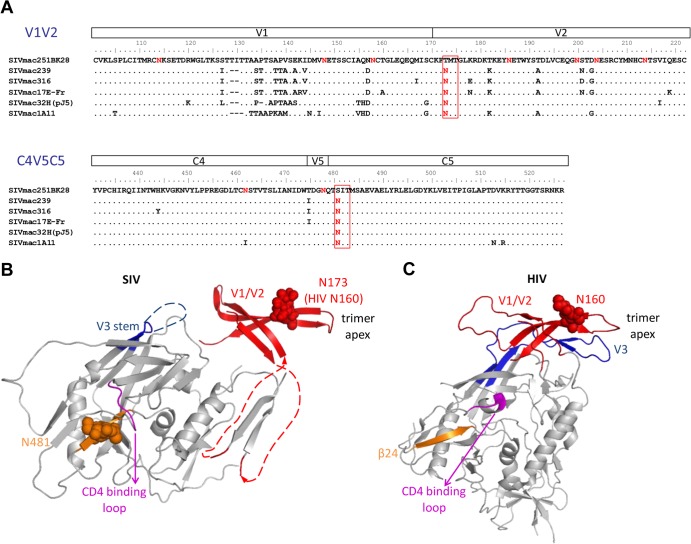
FIG 1.
N-linked glycosylation sites N173 and N481 are highly conserved among SIV strains but are missing in the SIVmac251BK28 clone. (A) Alignment of SIV gp120 sequences of the V1/V2 and the C4-V5-C5 regions shows that the N173 and N481 glycosylation sites are conserved in other well-studied SIV clones but are missing in SIVmac251BK28 (48). SIVmac239 and SIVmac32H (pJ5) are T-cell tropic (39, 95–97), whereas the other clones are macrophage tropic (6, 39, 48, 98–101). (B and C) N-linked glycosylation sites are shown in red, with N173 and N481 highlighted in red rectangles. The two N-linked glycosylation sites are mapped onto an unliganded SIV gp120 structure (41) (B) and an HIV gp120 structure from a soluble cleaved HIV Env trimer (44) (C), with the first sugar residues of the glycans and asparagine side chains shown as spheres. N173 and the V1/V2 region were substituted with a GAG linker in the original SIV gp120 structure. Here, we removed the GAG linker and modeled an HIV V1/V2 domain structure containing the analogous N160 glycan (red) (24), based on the orientation of V1/V2 in the HIV Env trimer structure. In the HIV Env trimer, V1/V2 forms a four-stranded β-sheet associated with V3 (blue) at the top of the Env trimer. The analogous N160 glycan is near the trimer apex. SIV N481 (orange) is at the β24 strand in the C5 region, in close proximity to the conserved GGDPE domain of the CD4 binding loop (magenta). The HIV Env protein shown here lacks the glycosylation site analogous to SIV N481. The β24 strand is shown in orange, as in the SIV gp120 structure.