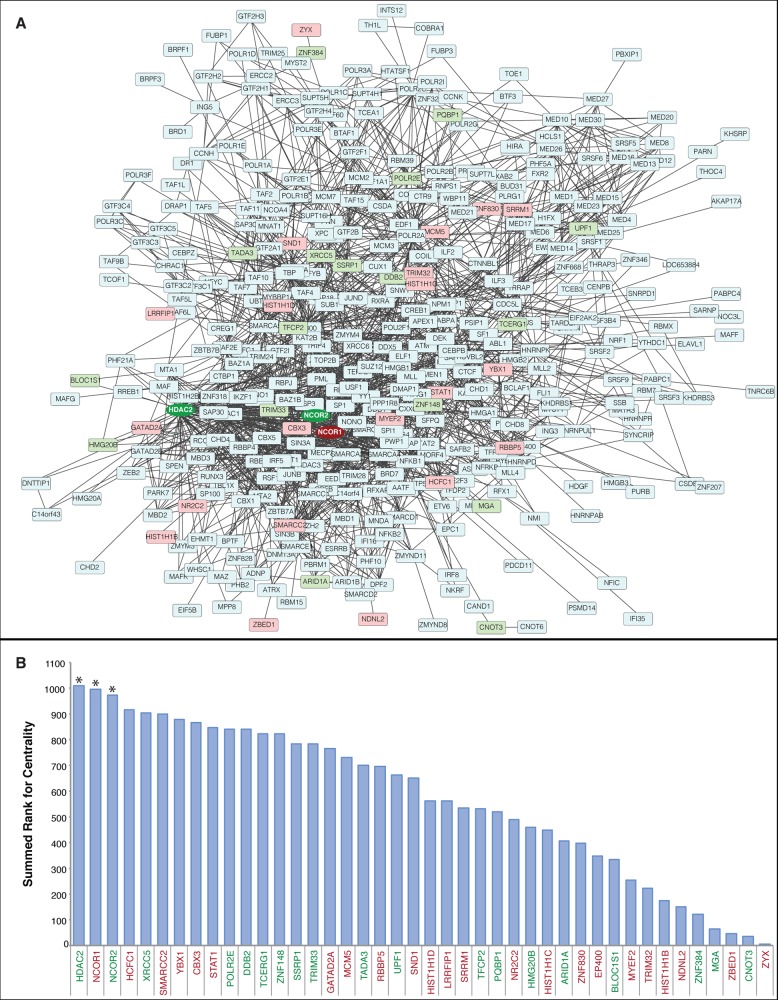
Figure 4.
Centrality analysis reveals critical nodes among interacting transcription regulators. (A) Cytoscape visualization of a STRING-generated network composed of experimentally verified protein–protein interactions among the transcription regulators identified in HIV-1 infected and uninfected MDM. Proteins in blue were identified, but not significantly altered. Proteins in red displayed increased expression in HIV-1-infected MDM, whereas proteins in green displayed decreased expression. The most central differentially expressed proteins (HDAC2, NCOR1, NCOR2) have darkened color and the node shapes are circular. (B) Centrality analysis using the node parameters of closeness, eccentricity, and radiality. Proteins were assigned a rank for each centrality measure, and the summed rank was used to sort proteins from higher centrality (left) to lower centrality (right). Differentially expressed proteins in the upper 10th percentile for summed rank are indicated with an asterisk (*). Red protein names indicate increased expression in HIV-1-infected MDM, whereas green protein names indicate decreased expression. The complete centrality analysis is provided in Supplementary data set 8.