Figure 4.
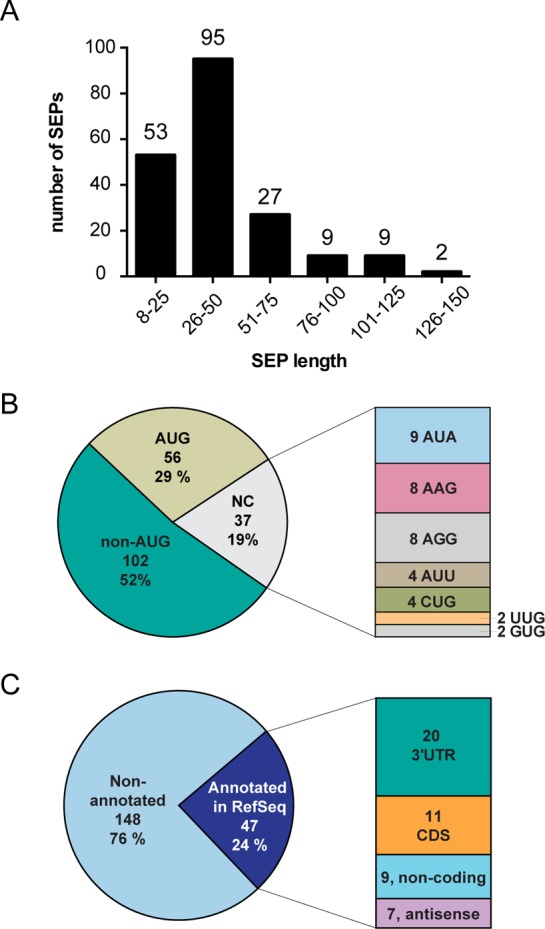
Overview of 195 novel SEPs identified in K562 cells. (A) Length of each SEP was determined using a defined set of criteria (see Methods), and the length distribution reveals that the majority (>90%) of SEPs discovered are between 8 and 100 amino acids. (B) SEPs utilize AUG, near cognate codons (i.e., one base away from AUG), and unknown codons to initiate translations. (C) SEPs are primarily derived from nonannoated RNAs (i.e., not found in RefSeq database), but RefSeq RNAs do account for the production of 24% of these SEPs. For the RefSeq-RNAs, the sORFs are found on coding RNAs at the 3′-UTR and CDS and on noncoding RNAs such as antisense RNAs and noncoding RNAs.
