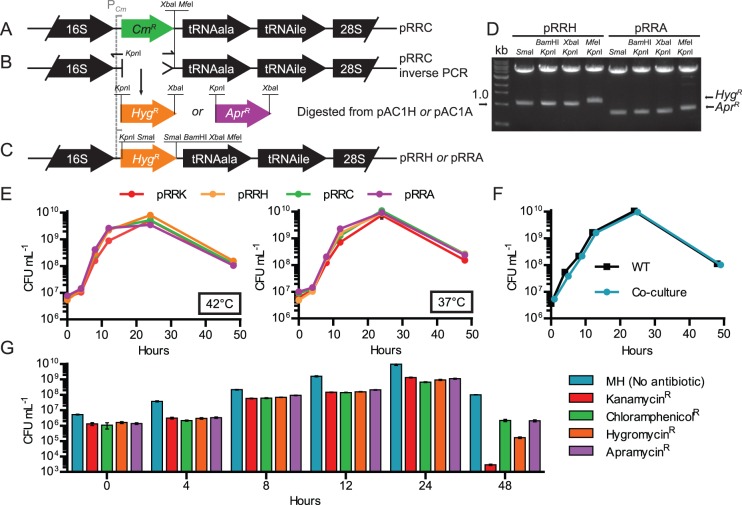
Figure 2. Adaptation of the pRRC gene delivery and expression system to harbor hygromycin B or apramycin resistance, and testing of genome-integrated markers for detrimental effects of resistance genes.
(A) Schematic of pRRC, which inserts into any of 3 rRNA clusters in the genome by homologous recombination. (B) Inverse PCR amplification of pRRC with primers 5705 (KpnI) and 5706 deleted the chloramphenicol resistance gene but conserved the Campylobacter-optimized cat promoter. (C) The inverse PCR product was digested with KpnI and XbaI, and ligated to similarly digested aph(7″) or aac(3)IV from pAC1H or pAC1A to create pRRH and pRRA respectively (only pRRH is shown). (D) Restriction digest analysis confirmed the function of all introduced sites. (E) The resistance markers from pRRK, pRRC, pRRH and pRRA were inserted into the C. jejuni 81–176 genome, and each resulting strain was analyzed for microaerobic growth and survival in shaken Mueller-Hinton (MH) broth by counting CFU over 48 hours at both 42°C (left panel) and 37°C (right panel). (F) To determine if the introduction of either marker contributed any fitness cost that could affect competitiveness against wild-type or the other marked strains, a competition assay was performed. Equal numbers of wild-type marked with hygromycin, apramycin, chloramphenicol and kanamycin resistance markers were co-cultured with unmarked wild-type in shaking MH broth at 37°C under microaerobic conditions. CFU were assessed by plating a dilution series on MH agar. (G) CFU were further assessed from the co-culture by plating on MH only (the total CFU, same data as in F) or MH supplemented with each antibiotic, representing the number of bacteria resistant to each antibiotic.