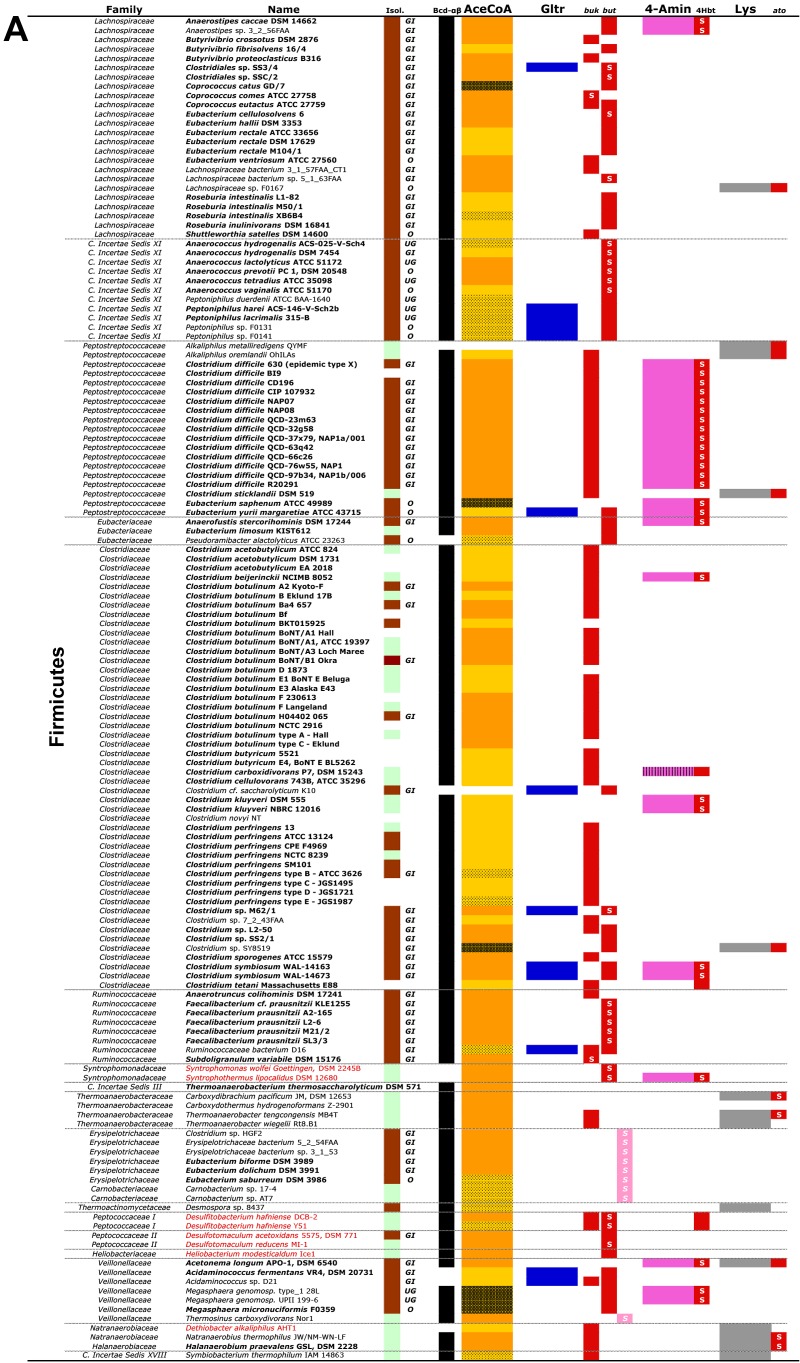
FIG 2 .
A list of all obtained candidate bacteria and their taxonomic classifications. Firmicutes are shown in panel A, whereas candidates associated with other phyla are displayed in panel B. Names in bold represent known butyrate-producing strains. Origins of isolates (Isol.), where brown refers to human/animal-associated strains (individual body sites of isolation are as follows: GI, gastrointestinal tract; UG, urogenital tract; O, oral tract) and green to environmental isolates, are given. Individual pathways with corresponding final genes are shown, namely, the acetyl-CoA pathway (AceCoA; orange-yellow) and the glutarate pathway (Gltr; blue) with but (encoding butyryl-CoA:acetate CoA transferase; red; light pink represents “atypical” transferases) and buk (butyrate kinase; red), as well as the 4-aminobutyrate pathway (4-Amin; pink) with the 4Hbt gene (butyryl-CoA:4-hydroxybutyrate CoA transferase; red) and the lysine pathway (Lys; grey) with ato (encoding butyryl-CoA:acetoacetate CoA transferase). Results of synteny analysis for genes of individual pathways are indicated (see key to color patterns at the bottom). Black cells in the column “Bcd-αβ” represent the presence of the butyryl-CoA dehydrogenase electron transfer protein complex, i.e., bcd is in synteny with the etf genes. Names in red indicate isolates that are reported to oxidize butyrate for growth. Actinob., Actinobacteria; Spro., Spirochaetes; The., Thermotogae; Bact., Bacteroidetes; C. Incertae Sedis, Clostridiales incertae sedis. For more explanation, see the text.