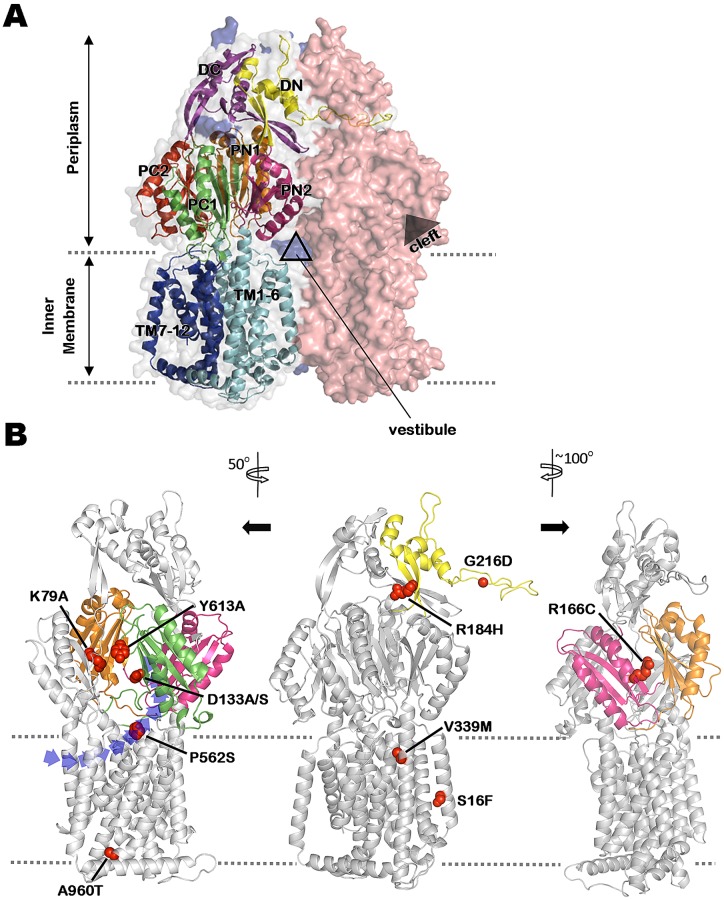
FIG 2 .
Mapping mutations impacting MexY-mediated antibiotic resistance on a 3-dimensional homology model of MexY. (A) An asymmetric trimer model of MexY constructed by homology modeling on the crystal structure of E. coli AcrB (PDB code 2HRT). Individual monomers are shown in space-fill formatting and are colored salmon red (loose/access conformation), gray (tight/binding conformation), and blue (open/extrusion conformation; mostly hidden behind the other 2 monomers). Structural details of the tight/binding monomer are shown in order to highlight the characteristic N- and C-terminal regions/subdomains of the docking domain (DN [yellow] and DC [purple], respectively), the porter domain (2 N-terminal [PN1, orange]/[PN2, pink] and 2 C-terminal [PC1, green]/[PC2, red] subdomains), and the transmembrane domain (transmembrane segments TM1-6 [light blue] and TM7-12 [dark blue], respectively) of RND transporters. The positions of the intermonomer vestibule and intramonomer cleft are also indicated. (B) Locations in the tight/binding MexY monomer of mutations (shown in space-fill formatting) that compromise MexY-mediated drug resistance. Relevant subdomains (as described for panel A) are highlighted in the corresponding colors. Structural models at left and right were rotated 50° counterclockwise and 100° clockwise, respectively, relative to the middle model in order to better illustrate the positions of various mutations. The dashed lines define the inner-membrane boundary. A cleft/opening, within which the putative proximal binding pocket occurs, is clearly seen in the structural model at left. A vestibule drug entry pathway is also highlighted in the leftmost structural model.