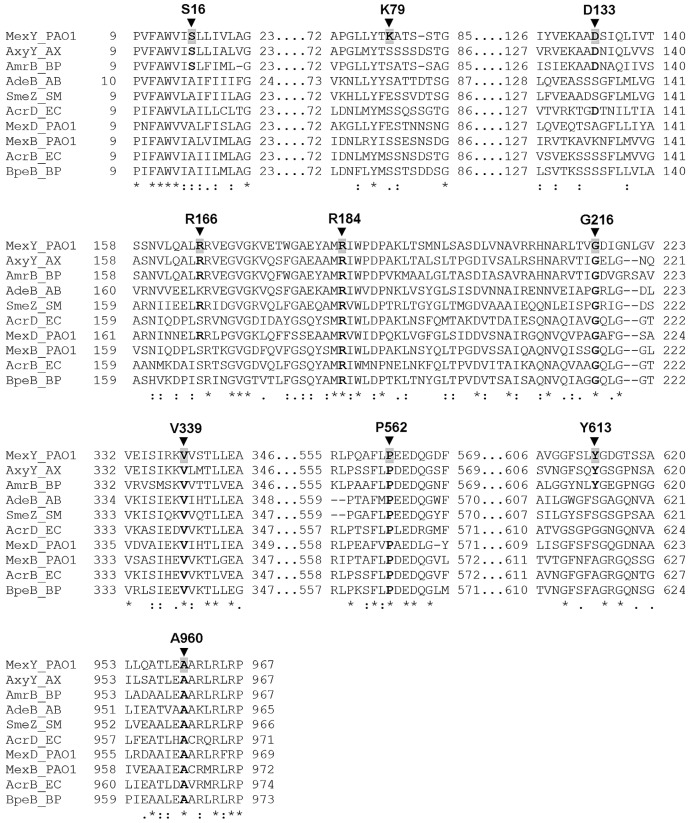
FIG 3 .
Segments of amino acid sequence alignment of MexY and selected RND homologues in P. aeruginosa and other Gram-negative bacteria. The amino acid sequences of MexY and the indicated RND homologues were aligned using the multiple-sequence-alignment program T-coffee (55). Only those aligned sequences that encompassed the MexY residues whose mutation compromised drug resistance (indicated with an arrowhead and labeled) are shown. Residues that are conserved in other RND homologues are highlighted in bold lettering. Amino acid sequences were obtained from the NCBI Protein database (GenBank accession no. NP_250708 [MexY_P. aeruginosa PAO1], EGP45231 [AxyY_Achromobacter xylosoxidans AXX-A], YP_108402 [AmrB_Burkholderia pseudomallei K96243], CAJ77844 [AdeB_Acinetobacter baumannii AYE], YP_001972001 [SmeZ_Stenotrophomonas maltophilia K279a], NP_416965 [AcrD_Escherichia coli K-12 MG1655], NP_253288 [MexD_P. aeruginosa PAO1], NP_249117 [MexB_P. aeruginosa PAO1], NP_414995 [AcrB_E. coli K-12 MG1655], and YP_006275722 [BpeB_B. pseudomallei 1026b]). MexY, AxyY, AmrB, AdeB, SmeZ, and AcrD have been linked to aminoglycoside resistance.