Abstract
Background & objectives:
Diagnosis of extrapulmonary tuberculosis (EPTB) is difficult using conventional diagnostic methods. This study was conducted to evaluate the use of polymerase chain reaction (PCR) in diagnosis of definitive and probable extrapulmonary tuberculosis patients, and to assess the performance of insertion sequence (IS) 6110 based PCR assay as compared to conventional culture by Lowenstein-Jensen (LJ) method for the diagnosis of EPTB.
Methods:
A total of 178 non repeated clinical specimens were collected from clinically suspected extrapulmonary tuberculosis patients. The specimens included 59 ascitic fluid, 54 pleural fluid, 25 cerebrospinal fluid (CSF), 12 fine needle aspiration (FNA), 8 urine, 7 pus, 6 synovial fluid, 2 skin tissue, one pericardial fluid, one liver abscess, one pancreatic cyst fluid, one omental biopsy and one semen sample. All these clinical samples were subjected to Ziehl-Neelsen staining (ZN) for acid fast bacilli (AFB) and culture on LJ medium. PCR was performed by targeting 123bp fragment of insertion sequence IS6110 of Mycobacterium tuberculosis (MTB).
Results:
Of the 178 specimens, 10 (5.61%) were ZN smear positive for AFB, six (3.37%) were L-J culture positive from 10 AFB smear positive cases and 48 (26.96%) were PCR IS 6110 positive for M. tuberculosis.
Interpretation & conclusions:
PCR using IS6110 primer was able to pick up more EPTB patients compared to conventional L-J culture method for detection of M. tuberculosis. False positive PCR IS6110 in three CSF samples may be due to latent TB infection which was limitation in this study.
Keywords: Acid fast bacilli, extra-pulmonary tuberculosis, IS6110 sequence, Lowenstein-Jensen medium, Mycobacterium tuberculosis, polymerase chain reaction
Tuberculosis (TB) is a major cause of morbidity and mortality worldwide infecting approximately one third of the population1. Tuberculosis spreads to almost every organ of the body and manifest clinically on the basis of localization of the infection. It is difficult to diagnose extrapulmonary tuberculosis (EPTB) by using the conventional diagnostic methods. Direct AFB (acid fast bacilli) smear and L-J (Lowenstein-Jensen) culture lack sensitivity for diagnosis of extrapulmonary tuberculosis and culture takes at least two weeks. Smear for AFB is reported to be positive in 10 to 37 per cent of patients and L-J culture is reported positive in variable proportion (12 to 80%) in different body fluids from suspected extrapulmonary tuberculosis cases2. The existing methodologies remain ineffective due to less number of mycobacteria and/or time consuming procedures3. Accurate and early diagnosis of TB is crucial for effective management and timely treatment. At present, nucleic acid amplification based assays are the most suitable choices for the identification of Mycobacterium tuberculosis in smear negative samples with high degree of sensitivity and specificity4,5. Several studies have been performed to detect M. tuberculosis in pulmonary and extrapulmonary clinical samples using PCR targeting different DNA sequences of M. tuberculosis6,7. The purpose of this study was to assess the utility of PCR in diagnosis of definitive and probable extrapulmonary tuberculosis patients, and to assess the performance of insertion sequence (IS) 6110 based PCR assay as compared to conventional culture by L-J method for the diagnosis of EPTB.
Material & Methods
All 178, clinically suspected extrapulmonary tuberculosis patients who were visiting inpatient and outpatient of various clinical departments of SRM Medical College Hospital & Research Centre, Kattankulathur, Kanchipuram District, Tamil Nadu, India, during the period of May 2008-May 2009, were included in the study. Written informed consent was obtained from each patient and the study protocol was approved by the Institutional Ethical Committee (IEC).
The following definitions were categorized based on the clinical profiles of the patients: (i) Definitive TB groups - Patients with AFB smear positive, L-J culture positive, histopathology positive (for relevant cases), Tuberculin test positive (10 mm or above), positive pulmonary findings in chest X-ray and previous history positive for TB; (ii) Probable TB groups - Patients with ambiguity in chest X-ray abnormalities, ultrasonagraphic (USG) findings, cytology, computerised tomography (CT) scan and cystoscopy; and (iii) Confirmed non TB groups.
Sterile body fluid samples (ascitic fluid, pleural fluid, CSF, synovial fluid, pericardial fluid and pancreatic cyst fluid) were centrifuged at 3000 g for 15 min. Pus specimens were decontaminated by Petroff`s method (4% NaOH) for 30 min8. Three consecutive early morning urine samples were collected and centrifuged at 3000 g for 15 min and the supernatant fluid was discarded. The deposit was decontaminated with 1 ml of 5 per cent H2SO4 for 15 min. Omental biopsy and skin tissue samples were grinded well with 5 ml of sterile distilled water. The specimens were centrifuged and the supernatant fluid was discarded. The deposit was decontaminated with 1 ml of 5 per cent H2SO4 for 15 min. One portion of all processed extrapulmonary clinical specimens were inoculated into a pair of L-J medium. Fine needle aspiration samples were directly inoculated in to a pair of L-J without decontamination. The second portions of all extrapulmonary clinical specimens were stored at -20°C in order to be used at a later stage for PCR work. The inoculated L-J media was examined every second day during the first week and weekly for up to 8 wk to monitor the presence of mycobacterial growth. Cultures grown were identified by standard morphological and biochemical tests.
Polymerase chain reaction
DNA extraction from extrapulmonary clinical specimens: An aliquot of centrifuged samples kept frozen at -20°C was used for PCR analysis.
Processing of tissue sample: Omental biopsy and skin tissue samples were finely chopped using a sterile scalpel and were homogenized manually in TE buffer (Tris- EDTA-10Mm Tris HCl, pH 8.0) until the solution turned out to be turbid. This was centrifuged at 11200 g for 20 min. FNA samples were directly utilized for DNA extraction. All other extrapulmonary clinical specimens were microcentrifuged at 11200 g for 5-10 min and then used for DNA extraction. A single portion of all extrapulmonary clinical specimens subjected with DNA extraction by standard (Cetyl trimethyl ammonium bromide) CTAB method9.
Amplification of mycobacterial DNA: A pair of oligonucleotide primers targeting insertion sequence 1S6110 of 123 bp length fragment specific for M. tuberculosis complex was used in this study: Forward Primer IS6110 a (5’ - CCT GCG AGC GTA GGC GTC GG -3’) and Reverse primer IS6110 b (5’ - CTC GTC CAG CGC CGC TTC GG - 3’) (Bangalore Geni, Bangalore, India). The IS6110 repetitive insertion sequence was designed for specific pair of primers to amplify 123bp as reported earlier10. Amplification was carried out in a final volume of 25 μl containing 10Mm Tris HCl (pH 8.0), 1.5 mM MgCl2, 200 μM of dNTPs, 20 Pico moles of each primer and 1.25 units of Taq polymerase (Biobasic, Canada). Five μl sample DNA was added to 25 μl of reaction mixture. Every batch of PCR had both negative and positive controls. The negative control had PCR grade water instead of the DNA sample and positive control had DNA of M. tuberculosis H37RV strain. Reagents were aliquoated and each aliquot was utilized only once.
Amplification protocol: The PCR amplification was done in thermal cycler (Applied biosystem Gene Amp PCR system 9700, Applied Biosystem, USA). Each cycle consisted of three steps comprising denaturation at 95°C for 5 min, followed by annealing of primers at 58°C for 30 sec, extension at 72°C for 30 sec with 35 cycles and a final extension at 72°C for 5 min.
Detection of amplified products: Amplified products were resolved by agarose gel electrophoresis (2%) at 80 volts for 40 min. Gel was stained with ethidium bromide (0.5 μg/ml) and viewed under UV transilluminator (VILBER -LOURMAT, France, TCP - 20.M).
Statistical analysis: Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the PCR results were evaluated against gold standard L-J culture method by using bivariate two by two tables (Binary classification method).
Results
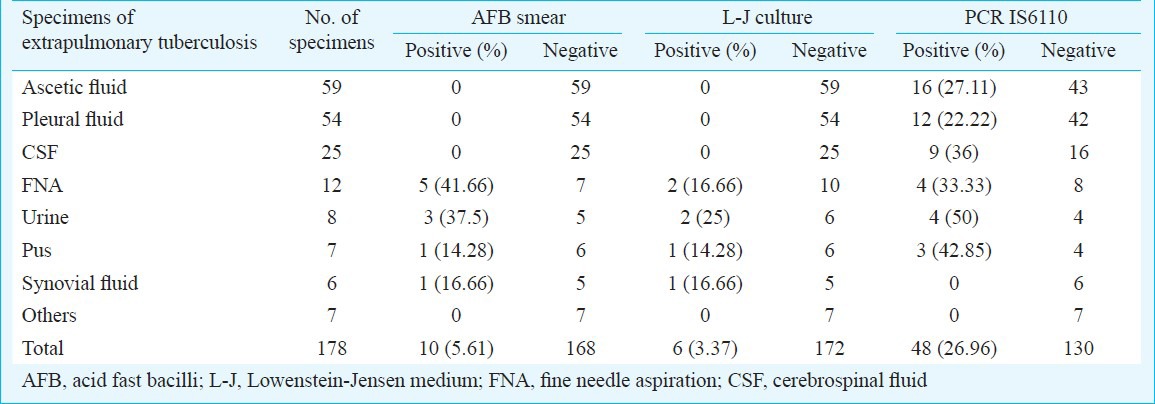
Among 178 suspected extrapulmonary tuberculosis patients, 36 (20.22%) were definitive extrapulmonary tuberculosis, 120 (67.4%) patients were probable extrapulmonary tuberculosis and 22 (12.35%) were non-TB groups. Ten (5.61%) samples were AFB positive by smear and 6 (3.37%) were L-J culture positive for
M. tuberculosis. DNA was detected by PCR in 48 samples (26.96%) which included 16 ascitic fluid samples, 12 pleural fluids, 9 CSF, 4 urine, 4 fine needle aspirations and 3 pus samples (Table I).
Table I.
AFB smear, LJ culture and PCR IS6110 positivity rate in various specimens of suspected extrapulmonary tuberculosis patients (N=178)
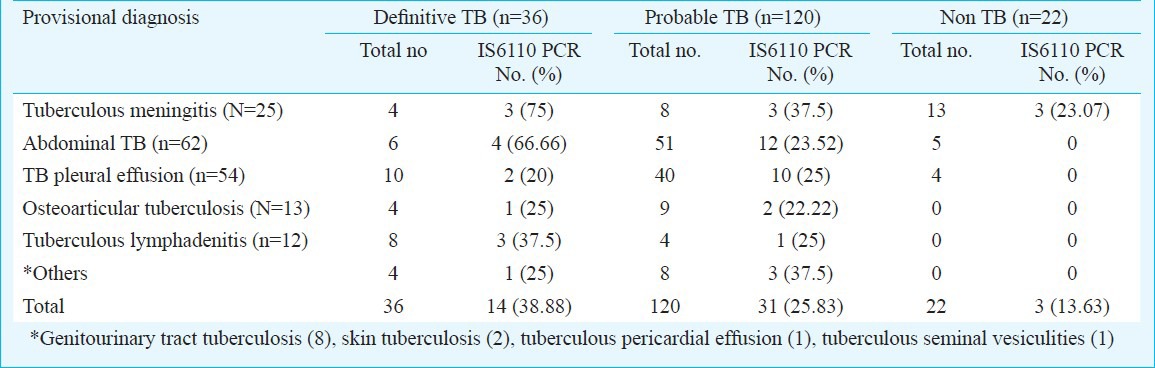
Among the definitive TB group, PCR IS6110 was positive in 14 of the 36 (38.88%) specimens. Whereas, in the 120 probable TB group, 31 (25.83%) specimens were positive by PCR IS6110 and in the 22 non-TB group three samples (13.63%) were PCR IS6110 positive (Table II).
Table II.
PCR positivity with respect to clinical presentations of extrapulmonary (EP) TB cases (N=178)
Of the 22 confirmed non TB samples, four were pleural effusions and five were peritoneal effusions. All nine effusion fluids were proved to be malignant by cytological analysis. Ten CSF samples were finally diagnosed to be viral meningoencephalopathy. Three CSF samples from clinically suspected pyogenic meningitis were found to be IS6110 PCR positive. These patient responded to treatment and recovered from the symptoms. Hence, these three PCR positives were considered as false positives.
There was no amplification in six AFB smear positive (1 synovial fluid, 2 urine and 3 FNA) samples. These six samples were tested for the presence of substances inhibiting Taq polymerase by spiking duplicate samples with M. tuberculosis DNA and found to be negative for the presence of inhibitors. The two samples of synovial fluid and lymph node aspirate where AFB smear and L-J culture were positive but PCR was negative, could be due to presence of PCR inhibiting substances in the sample.
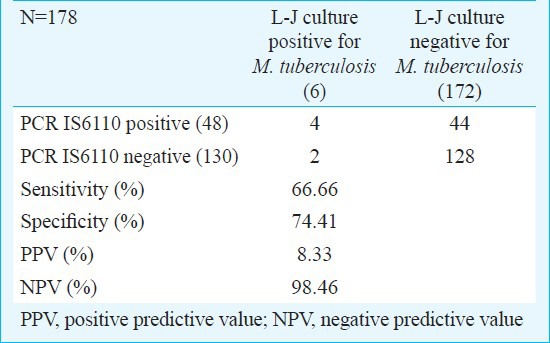
IS6110 PCR has shown 66.66 per cent sensitivity (with 95% confidential interval (CI) 24.1; 94) and 74.41 per cent specificity (with 95% CI, 67.1; 80.6). Overall positive and negative predictive value of IS6110 PCR was observed as 8.33 per cent (with 95% CI, 2.7; 20.8) and 98.46 per cent (with 95% CI, 93.9; 99.7) (Table III).
Table III.
Comparison of PCR IS6110 results with conventional Lowenstein-Jensen (L-J) medium
Discussion
Extrapulmonary tuberculosis is a significant health dilemma in both developed and developing countries11. A high degree of suspicion aided by intensive investigations is important in the diagnosis of the extrapulmonary tuberculosis. The role of routine diagnostic methods are of little value. The diagnosis of EPTB in different clinical presentations still remains a challenge. Conventional methods have a very low sensitivity in the diagnosis of extrapulmonary clinical specimens, perhaps due to unequal distribution of acid fast bacilli in large volumes of fluids. For the smear to be AFB positive, the sample should contain at least 10000 bacilli/ml. L-J culture is still considered to be the gold standard, but 10-100 viable bacilli are mandatory for culture positivity. Moreover, long period of time (of about 6-8 wk) is required for positive reports, hence most clinical and therapeutic decisions cannot be made12. In addition, the histopathological findings suggestive of granulomatous infection may encompass many differential diagnoses2.
Several M. tuberculosis specific target DNA sequences have been tried so far for the diagnosis of pulmonary and extrapulmonary tuberculosis by PCR and various other genotypic methods13,14,15. These include gene coding for the 65 kDa heat shock protein (HSP), IS 6110 insertion sequences, gene coding for 38 kDa, 85B antigen and 16S rRNA. IS6110 as target sequence for detection of mycobacterial DNA from extrapulmonary clinical samples showed wide variation in specificity and sensitivity16,17,18,19,20. IS6110 is a long 1191 bp repetitive insertion sequence that is usually present 6-20 times in the M. tuberculosis complex genome than other repetitive sequence21. Negi et al22 reported the amplification the same 123bp fragement targeted IS6110 in their study. Among their samples, 83 per cent were PCR positive. Ogusku et al23 showed 92.1 per cent of their samples to be IS6110 positive compared with 65 kDa, 38 kDa and MPB 64 specific primers. Amin et al24 reported 38.6 per cent positivity rate in pus, 42.1 per cent in CSF and 46.6 per cent in urine samples. In comparison, our study revealed 50 per cent positivity in urine, 42.85 per cent in pus and 36 per cent in CSF samples. An earlier Indian study25 also reported 63 per cent positivity by PCR using IS 6110 element in specimens of EPTB. Most of the studies which used IS6110 based PCR, reported 90 per cent sensitivity in CSF, pleural fluid, ascitic fluid and other extrapulmonary specimens25. Tiwari et al14 showed 62 per cent total positivity rate among EPTB samples and detection of M. tuberculosis DNA in 57 per cent of AFB smear negative EPTB samples. Our study showed a low number of PCR positives among the 154 body fluid samples, i.e. 27 per cent in ascetic fluid, 22 per cent pleural fluids and 36 per cent CSF samples.
IS6110 is specific for M. tuberculosis complex and generally occurs in 1-20 copies per cell, which are dispersed in the M. tuberculosis genome and it an ideal target for amplification, one locus, the direct repeat region, has on high frequency of carriage of IS6110 and has been proposed as a “hot spot” for integration of this element, although most of the copies are located at a single site26. These insertion elements are present in multiple copies on the genome of M. tuberculosis, with 16 copies of IS 6110, 6 copies of IS1081 and 2 copies each of IS154727. The variable copy number of IS6110 among different strains of the tubercle bacilli has led to its extensive use as a genetic marker to investigate the epidemiology of tuberculosis28.
The common problem raised during the PCR assays is the high risk of false positive results due to common laboratory contamination or presence of killed or dormant bacilli in the patient specimens29,30. Proper control checks and good laboratory practice can minimize the chances of false positive results. There are several other possible reasons for false negativity viz., the paucibacillary nature of the disease, possible hypersensitivity mechanisms, or the availability of only one small amount or volume of sample after it was distributed for various microbiological, pathological and biochemical investigations. The drawback with PCR assay is that it is not able to differentiate live from dead organisms. The advantages of IS6110 PCR are that it is very rapid, easy to perform method and result can be issued for early treatment and to prevent further transmission of tuberculosis infection. Further, IS 6110 PCR test proved to be more sensitive even when both smear examination and culture results were considered in conjunction.
In conclusion, the present study revealed that PCR using IS6110 as primer could detect more number of positives in extrapulmonary tuberculosis compared with conventional methods. Though PCR using IS6110 showed increased sensitivity compared to conventional L-J culture, and was a rapid method, the false positivity was a limitation.
Acknowledgment
Financial assistance in the form of a University Research Fellowship (URF) provided by the SRM Medical College Hospital and Research Centre, SRM University, Kattankulathur, Kanchipuram district, to the first author (VM) is acknowledged. Authors thank to all clinicians of SRM Medical College Hospital and Research Centre for providing clinical samples and the Director, National Institute for Research in Tuberculosis (Formerly Tuberculosis Research Centre), Indian Council of Medical Research, Chennai for permitting the use of Molecular Biology laboratory during this study.
References
- 1.Geneva, Switzerland: World Health Organization; 2002. World Health Organization. Global tuberculosis control. WHO Report 2002. WHO/CDS/TB/2002.295. [Google Scholar]
- 2.Sharma SK, Mohan A. Extrapulmonary tuberculosis. Indian J Med Res. 2004;120:316–53. [PubMed] [Google Scholar]
- 3.Styrt BA, Shinnick TM, Ridderhof JC, Crawford JT, Tenover FC. Turnaround times for mycobacterial cultures. J Clin Microbiol. 1997;35:1041–2. doi: 10.1128/jcm.35.4.1041-1042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soni H, Musser JM. Molecular diagnosis of mycobacteria. Clin Chem. 2001;47:809–14. [PubMed] [Google Scholar]
- 5.Hale YM, Pfyffer GE, Salfinger M. Laboratory diagnosis of mycobacterial infections: new tools and lessons learned. Clin Infect Dis. 2001;33:834–46. doi: 10.1086/322607. [DOI] [PubMed] [Google Scholar]
- 6.Montenegro SH, Gilman RH, Sheen P, Cama R, Caviedes L, Hopper T, et al. Improved detection of M. tuberculosis in Peruvian Children by use of heminested IS 6110 PCR assay. Clin Infect Dis. 2003;36:16–23. doi: 10.1086/344900. [DOI] [PubMed] [Google Scholar]
- 7.Bennedsen J, Thomsen VO, Pfyffer GE, Funke G, Feldmann K, Beneke A, et al. Utility of PCR in diagnosing pulmonary tuberculosis. J Clin Microb. 1996;34:1407–11. doi: 10.1128/jcm.34.6.1407-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts GD. Mycobacteria and Norcardia. In: Washington JA II, editor. Laboratory procedures in clinical microbiology. New York: Springer Verlag; 1981. pp. 365–406. [Google Scholar]
- 9.Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43:2996–7. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenach KD, Cave MD, Bates JH, Crawford JT. Polymerase chain reaction amplification of a repetitive DNA sequences specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161:977–81. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 11.Cailhol J, Decludt B, Che D. Scoiodemographic factors that contribute to the development of extrapulmonary tuberculosis were identified. J Clin Epidemiol. 2005;58:1066–71. doi: 10.1016/j.jclinepi.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Prasad R, Lath SK, Mukerji PK, Agrawal SK, Srivastava R. Clinical utility of polymerase chain reaction in patients of pulmonary tuberculosis. Indian J Tuberc. 2001;48:135. [Google Scholar]
- 13.Pao CC, Yen TS, You JB, Maa JS, Fiss EH, Chang CH. Detection and identification of Mycobacterium tuberculosis by DNA amplification. J Clin Microbiol. 1990;28:1877–80. doi: 10.1128/jcm.28.9.1877-1880.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiwari V, Jain A, Verma RK. Applification of enzyme amplified mycobacterial DNA detection in the diagnosis of pulmonary and extrapulmonary tuberculosis. Indian J Med Res. 2003;118:224–8. [PubMed] [Google Scholar]
- 15.Miyazaki Y, Konga H, Kohno S, Kaku M. Nested polymerase chain reaction for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1993;31:2228–32. doi: 10.1128/jcm.31.8.2228-2232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger EC. Detection and identification of mycobacteria by amplification of r RNA. J Clin Microbiol. 1990;28:1751–9. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fries JW, Patel RJ, Piessens WF, Wirth DF. Detection of untreated mycobacteria by using polymerase chain reaction and specific DNA probes. J Clin Microbiol. 1991;29:1744–7. doi: 10.1128/jcm.29.8.1744-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneko K, Onodera O, Miyatake T, Tsuji S. Rapid diagnosis of tuberculous meningitis by polymerase chain reaction (PCR) Neurology. 1990;40:1617–8. doi: 10.1212/wnl.40.10.1617. [DOI] [PubMed] [Google Scholar]
- 19.Kolk AH, Schuitema AR, Kuijper S, van Leeuwen J, Hermans PW, van Embden JD, et al. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a nonradioactive detection system. J Clin Microbial. 1992;30:2567–75. doi: 10.1128/jcm.30.10.2567-2575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierre C, Lecossier D, Boussougant Y, Bocart D, Joly V, Yeni P, et al. Use of a re-amplification protocol improves sensitivity of detection of Mycobacterium tuberculosis in clinical specimens by amplification of DNA. J Clin Microbiol. 1991;29:712–7. doi: 10.1128/jcm.29.4.712-717.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenach KD, Cave MD, Bates JH, Crawford JT. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161:977–81. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 22.Negi SS, Anand R, Pasha ST, Gupta S, Blasir SF, Khare S, et al. Diagnostic potential of IS6110, 38 kda, 65 kda and 85B sequence based polymerase chain reaction in the diagnosis of Mycobacterium tuberculosis in clinical specimens. Indian J Med Microbiol. 2007;25:43–9. doi: 10.4103/0255-0857.31061. [DOI] [PubMed] [Google Scholar]
- 23.Ogusuk MM, Salem JI. Analysis of different primers used in the PCR method: diagnosis of tuberculosis in the state of Amazonas. Brazil J Bras Pnemol. 2004;30:343–9. [Google Scholar]
- 24.Amin I, Idress M, Awan Z, Shahid M, Afzal S, Hussain A. PCR could be a method of choice for identification of both pulmonary and extrapulmonary tuberculosis. BMC Res Notes. 2011;4:332. doi: 10.1186/1756-0500-4-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayanan S, Parandaman V, Narayanan PR, Venkatesan P, Girish C, Mahadevan S, et al. Evaluation of PCR using TRC (4) and IS6110 primers in detection of tuberculous meningitis. J Clin Microbiol. 2001;39:2006–8. doi: 10.1128/JCM.39.5.2006-2008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strains identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for standardized methodology. J Clin Microbiol. 1993;31:406–9. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 28.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, et al. The epidemiology of tuberculosis in san Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–9. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 29.Beige J, Lokies J, Schaberg T, Finckh M, Fischer M, Mauch H, et al. Clinical evaluation of a Mycobacterium tuberculosis PCR assay. J Clin Microbiol. 1995;33:90–5. doi: 10.1128/jcm.33.1.90-95.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connelly Smith K, Starke JR, Eisenach K, Ong LT, Denby M. Detection of Mycobacterium tuberculosis in clinical specimens from children using polymerase chain reaction. Pediatrics. 1996;97:155–66. [PubMed] [Google Scholar]


