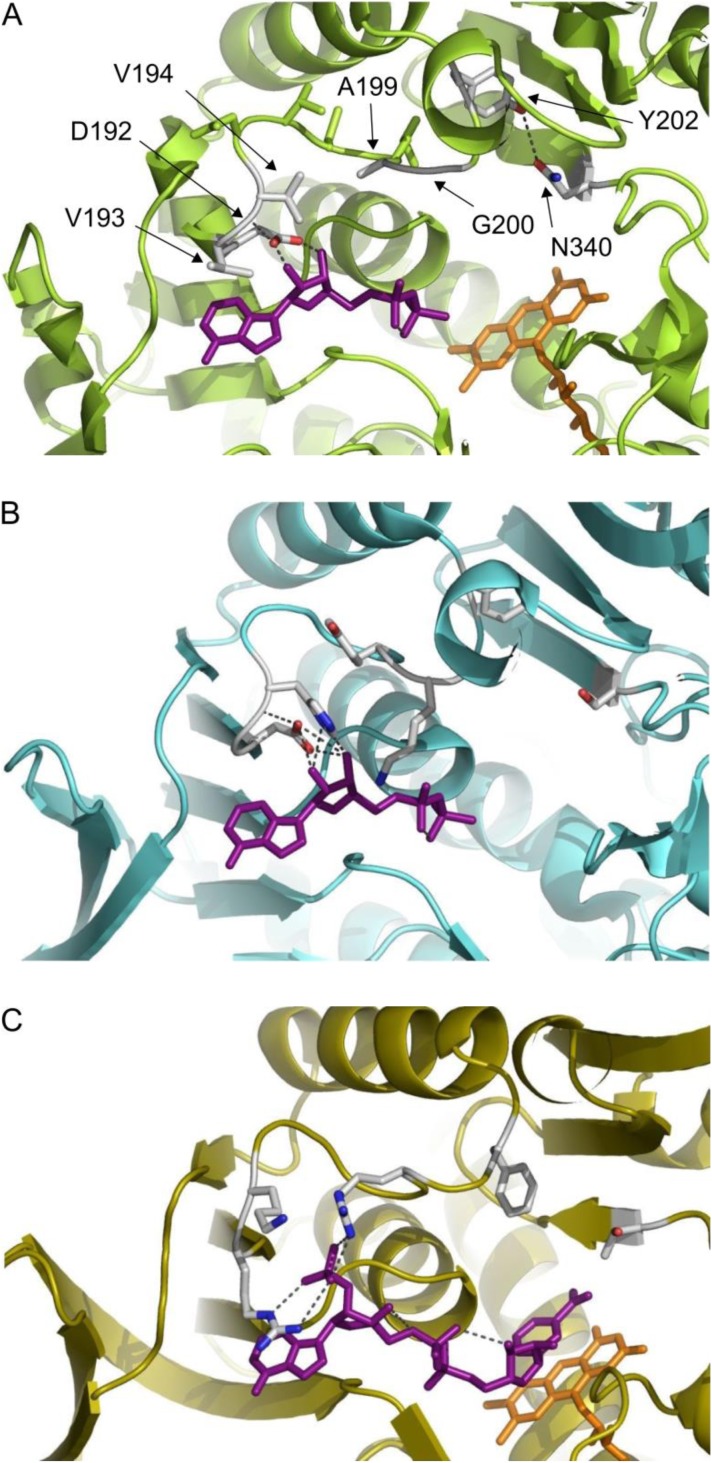
Figure 2.
Cofactor binding sites of S. mutans NADH oxidase 2, L. sanfranciscensis NAD(P)H oxidase and E. coli Glutathione reductase.
Panel A: Swiss-model for S. mutans NOX 2, based on S. pyogenes NOX structure (77.5% identity, pdb 2BC0), ADP from aligned LsNOX structure pdb 2CDU, FAD from pdb 2BC0; panel B: L. sanfranciscensis NAD(P)H crystal structure pdb 2CDU with ADP bound [60]; panel C: E. coli Glutathione reductase crystal structure pdb 1GET [61], images were generated with PyMOL (Schrodinger Inc.)