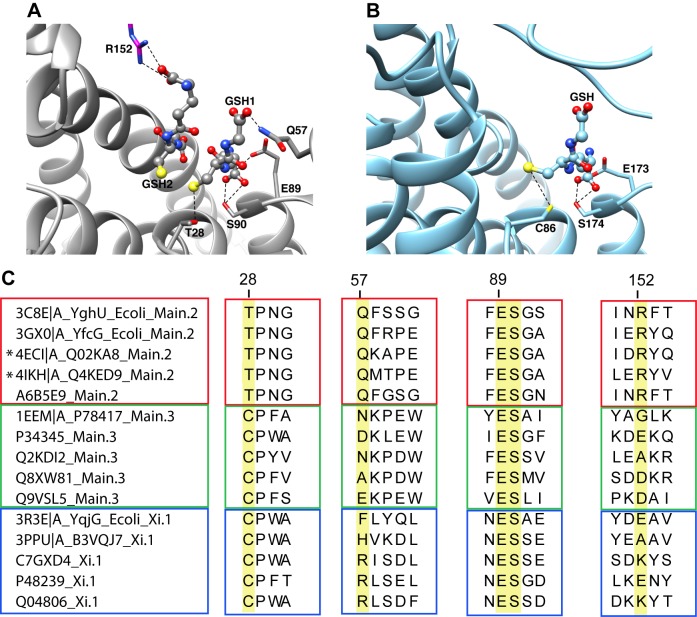
Figure 9. Comparison of the active site region from divergent subgroups with DSBR activity reveals some commonalities.
(A) Structure of Q4KED9 (UniProt accession) from Pseudomonas fluorescens (PDB ID 4IKH, subgroup Main.2), one of the new structures from this work, showing two molecules of glutathione bound in the active site. The interactions between the bound ligands and the side chains of Thr28, Gln57, Glu89, Ser90, and Arg152 of the other subunit (magenta) are shown. (B) Structure of B3VQJ7 from Phanerochaete chrysosporium (PDB ID 3PPU, subgroup Xi.1) with one molecule of glutathione bound. The corresponding interactions between glutathione and the side chains of Cys86, Glu173, and Ser174 are shown. (C) Summary of sequence motifs from the structure-guided alignment showing the sequence context for the residues highlighted in 7A and 7B from several divergent subgroups: Main.2 (red box), Main.3 (green box), and Xi.1 (blue box). All the proteins shown have experimental evidence for DSBR activity. UniProt entries and available PDB IDs are given on the left side of the sequences. Highlighted in yellow are the aligned positions of the residues that have notable interactions with the bound ligand as described in the text (numbered according to 4IKH/Q4KED9). New structures generated for this work are indicated with an asterisk.