Abstract
Insufficient epigenetic reprogramming of donor nuclei is believed to be one of the most important causes of low development efficiency of mammalian somatic cell nuclear transfer (SCNT). Previous studies have shown that both the in vitro and in vivo development of mouse SCNT embryos could be increased significantly by treatment with various histone deacetylase inhibitors (HDACi), including Trichostatin A, Scriptaid, and m-carboxycinnamic acid bishydroxamide (CBHA), in which only the effect of CBHA has not yet been tested in other species. In this paper we examine the effect of CBHA treatment on the development of porcine SCNT embryos. We have discovered the optimum dosage and time for CBHA treatment: incubating SCNT embryos with 2 μmol/L CBHA for 24 h after activation could increase the blastocyst rate from 12.7% to 26.5%. Immunofluorescence results showed that the level of acetylation at histone 3 lysine 9 (AcH3K9), acetylation at histone 3 lysine 18 (AcH3K18), and acetylation at histone 4 lysine 16 (AcH4K16) was raised after CBHA treatment. Meanwhile, CBHA treatment improved the expression of development relating genes such as pou5f1, cdx2, and the imprinted genes like igf2. Despite these promising in vitro results and histone reprogramming, the full term development was not significantly increased after treatment. In conclusion, CBHA improves the in vitro development of pig SCNT embryos, increases the global histone acetylation and corrects the expression of some developmentally important genes at early stages. As in mouse SCNT, we have shown that nuclear epigenetic reprogramming in pig early SCNT embryos can be modified by CBHA treatment.
Electronic supplementary material
The online version of this article (doi:10.1007/s13238-014-0034-3) contains supplementary material, which is available to authorized users.
Keywords: swine, nuclear transfer, epigenetic reprogramming, histone deacetylase inhibitor
Introduction
Somatic cell nuclear transfer (SCNT) has been a successful technology to derive cloned animals in many species (Wilmut et al., 1997; Kato et al., 1998; Wakayama et al., 1998; Baguisi et al., 1999; Onishi et al., 2000; Chesne et al., 2002; Shin et al., 2002; Galli et al., 2003; Woods et al., 2003; Zhou et al., 2003; Lee et al., 2005; Li et al, 2006). It is a valuable tool in fundamental research, biomedical and reproduction fields (Verma et al., 2011; Bendixen et al., 2013). As pigs share much similar physiological characteristics with human beings, it is a potential animal model for human disease and xenotransplantation (Gock et al., 2011; Ekser et al., 2012; Luo et al., 2012). SCNT is a practical way to produce targeted genetic modification in pigs.
However, like in other animals, SCNT in pigs is inefficient, due to the low cloning efficiency, fetus abnormality, and placenta deficiency (Polejaeva et al., 2000; Walker et al., 2002). It is widely believed that the crucial cause of poor developmental capacity in nuclear transfer embryos is the aberrant nuclear reprogramming. There is a close relationship between embryo reprogramming and epigenetic modification. DNA methylation and histone acetylation are the main modification patterns in epigenetic reprogramming. Compared with normal fertilized zygotes, in SCNT embryos nuclear, reprogramming has to occur within a short time span, so a more “open” chromatin configuration may be beneficial to SCNT embryos (William et al., 2001; Whitworth et al., 2010; Ogura et al., 2013). Histone deacetylase inhibitor (HDACi), has been proved to increase the acetylation level in somatic cells or in nuclear transfer embryos, as it makes the chromatin more flexible and allowing the combination of transcriptional factors (Kishigami et al., 2006; Lager et al., 2008; Dai et al., 2010).
In pigs, some histone deacetylase inhibitors, such as Trichostatin (TSA), Scriptaid, and Valproic acid (VPA), have been proved to increase the in vitro development of SCNT embryos (Li et al., 2008; Miyoshi et al., 2010; Zhao et al., 2010; Kim et al., 2011). Our previous study has shown that m-carboxycinnamic acid bishydroxamide (CBHA), a member of type II HDACi, could improve the in vitro and full term development of mouse SCNT embryos and improve the cell line establishment efficiency of embryonic stem cells (ES) derived from SCNT embryos (SCNT-ESCs) (Dai et al., 2010). However, CBHA has not yet been studied in domestic animals. In this study we use this HDACi to treat porcine nuclear transfer embryos to investigate whether it affects the developmental efficiency in pigs.
We have examined the optimum treatment program of CBHA on pig nuclear transfer embryos and observed the development rates in various treated and non-treated groups. To further explore how it improves the development of porcine nuclear transfer embryos, we detected the global acetylation levels of Histone 3 at Lysine 9 (AcH3K9), Histone 3 at Lysine 18 (AcH3K18), and Histone 4 at Lysine 16 (AcH4K16) using immunofluorescence in the CBHA treated embryos and control embryos. Then we analyzed the effect of CBHA treatment on the transcription level of some development-related genes, histone deacetylase genes, and imprinted genes using quantitative PCR. Finally, we detected the full term developmental efficiency of CBHA treated and non-treated embryos.
Results
In vitro development of porcine SCNT embryos are improved by CBHA treatment
To test whether CBHA could improve the early development of porcine SCNT embryos, we treated the SCNT embryos with different concentrations of CBHA and calculated the developmental rates from 2-cell to blastocyst stage (Table 1). We found that proper CBHA treatment had no effect on the cleavage rates of porcine SCNT embryos, but could significantly increase the in vitro development rate to blastocyst stage. The optimum treatment concentration of CBHA was 2 μmol/L, while 200 μmol/L or higher concentrations were detrimental to embryos. After treatment with 2 μmol/L CBHA for 24 h, the blastocyst ratio of CBHA-SCNT embryos was two folds (26.5%) of the embryos in control group (12.7%).
Table 1.
Pre-implantation development of pig SCNT embryos under treatment with different CBHA concentration
| CBHA Treatment (μmol/h) | No. of reconstructed embryos | No. of 2-cell (% reconstructed) | No. of blastocysts (% 2-cell) | No. of blastocysts (% reconstructed) |
|---|---|---|---|---|
| 0 | 212 | 132 (62.3) | 27 (20.5) | 27 (12.7 ± 5.7)a |
| 0.02/24 | 58 | 40 (69.0) | 8 (20.0) | 8 (13.8 ± 1.4)a |
| 0.2/24 | 123 | 75 (61.0) | 25 (33.3) | 25 (20.3 ± 9.0)b |
| 2/24 | 211 | 117 (55.4) | 56 (47.9) | 56 (26.5 ± 12.3)c |
| 10/24 | 90 | 60 (66.7) | 17 (28.3) | 17 (18.9 ± 17.9)b |
| 20/24 | 213 | 110 (51.6) | 35 (31.8) | 35 (16.4 ± 10.7)b |
| 50/24 | 90 | 35 (38.9) | 12 (34.3) | 12 (13.3 ± 3.8)a |
| 200/24 | 65 | 1 (1.5) | 0 (–) | 0 (–) |
| 400/24 | 65 | 0 (–) | 0 (–) | 0 (–) |
Significant χ2 comparisons: a versus c, P < 0.05; a versus b, P > 0.05; b versus c, P > 0.05
Next, to optimize the protocol for CBHA treatment, we examined the effect of different CBHA incubation times on the blastocyst ratios of SCNT embryos. The SCNT embryos were treated with 2 μmol/L or 10 μmol/L CBHA for 16 h, 20 h, and 24 h, respectively. There was no significant difference among the three groups, but the treatment of 2 μmol/L, 24 h gave the best blastocyst ratio (Table 2).
Table 2.
Effect of time duration of CBHA treatment on the pre-implantation development of SCNT embryos
| Concentration of CBHA (μmol /L) | Time duration (h) | No. of reconstructed embryos | No. of 2-cell (% reconstructed) | No. of blastocysts (% 2-cell) | No. of blastocysts (% reconstructed) |
|---|---|---|---|---|---|
| 2 | 16 | 77 | 40 (52.0) | 11 (27.5) | 11 (14.3 ± 12.5) |
| 20 | 78 | 44 (56.4) | 17 (38.6) | 17 (21.8 ± 3.8) | |
| 24 | 77 | 34 (44.2) | 16 (47.1) | 16 (20.8 ± 7.2) | |
| 20 | 16 | 73 | 34 (46.6) | 13 (38.2) | 13 (17.8 ± 13.2) |
| 20 | 80 | 38 (47.5) | 13 (34.2) | 13 (16.3 ± 13.9) | |
| 24 | 78 | 36 (45.2) | 10 (27.8) | 10 (12.8 ± 4.0) | |
| 0 | – | 80 | 33 (41.3) | 14 (42.4) | 14 (17.5 ± 6.7) |
Blastocyst quality was improved after CBHA treatment
To unravel the improvement effects of CBHA treatment on SCNT embryo development, we examined the blastocyst cell number in two groups (Fig. 1). Though the cell numbers in the treated group were higher than the non-treated group, there was no significant difference between the two groups (34 vs. 32). Cell numbers from both groups were lower than their in vivo and in vitro fertilized counterparts.
Figure 1.
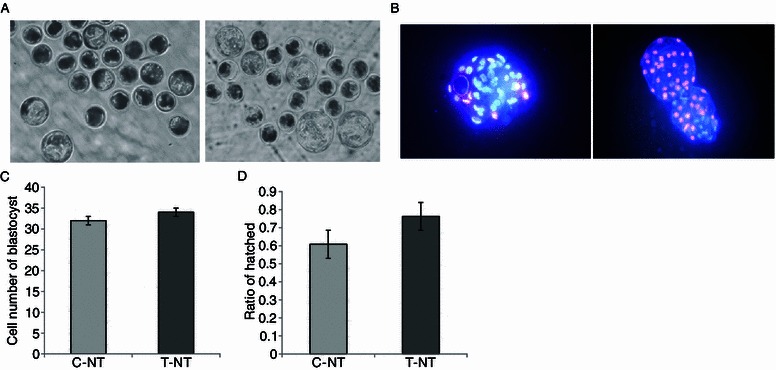
Effect of CBHA on hatching rate and cell number of SCNT blastocysts. (A) Representative photographs of pig blastocysts. Day 7 blastocysts developed from 0 μmol/L CBHA treated SCNT embryos (C-NT group) and 2 μmol/L CBHA treated SCNTembryos (T-NT group). Original magnification was 100×. (B) Fluorescent photographs of pig blastocysts stained with Hoechst. Original magnification was 200×. (C) Statistics of the number of cells in blastocyst from 0 μmol/L CBHA treated SCNT embryos (C-NT group) and 2 μmol/L treated embryos (T-NT group). (D) Ratio of hatched blastocysts from 0 μmol/L CBHA treated SCNT embryos (C-NT group) and 2 μmol/L treated embryos (T-NT group)
Embryo implantation and further development are largely related to the blastocyst-hatching rate and the communication between embryo and the uterus. Hence we examined the blastocyst-hatching rate in the two groups. The hatched blastocysts were observed under a stereomicroscope at 168 h after activation (Fig. 1). Although the hatching rate in the CBHA treated group (76.25%) was much higher than those in the control group (67%), there was no significant difference between these two groups. Nevertheless, we found that embryos treated with CBHA could reach the blastocyst stage much earlier than those in the control groups, indicating that at the developmental phase they are more like the in vivo embryos, who reach the blastocyst stage on about day 5.
CBHA treatment increases global histone acetylation levels of pig SCNT embryos
As shown in embryos, the global histone acetylation level is closely related to the reprogramming state of chromatin. Meanwhile, we showed that an open, decondensed chromatin state was relevant to the pluripotency of iPSCs (induced pluripotent stem cells) and ESCs (embryonic stem cells). We further examined the embryo histone acetylation levels by assaying three epigenetic markers: AcH3K9, AcH3K18, and AcH4K16 at 1-cell, 2-cell, and blastocyst stages. We found that CBHA treatment increased the level of AcH3K9, AcH3K18, and AcH4K16 at 1-cell (Fig. 2) and 2-cell stages (Fig. 3). However, apart from AcH3K18, at the blastocyst stage there were no differences between the control and experiment groups (Fig. 4).
Figure 2.
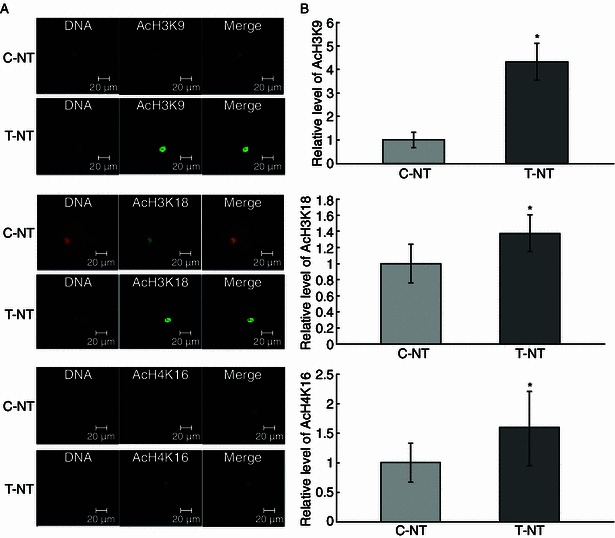
The global AcH3K9, AcH3K18, and AcH4K16 levels of 1-cell stage embryos. (A) Staining of AcH3K9, AcH3K18, and AcH4K16 (green) in non-treated control SCNT embryos (C-NT), and 2 μmol/L CBHA treated embryos (T-NT) at 1-cell stage. Each sample was counterstained with DAPI to visualize DNA (red). Original magnification was 200×. (B) Quantification of AcH3K9 and AcH3K18 signal intensities in C-NT (gray bars) and T-NT (black bars) embryos. Labeling intensity was expressed relative to that of the C-NT embryos (set as 100%). *P < 0.05. The experiments were replicated 3 times. n = 8–10 per group
Figure 3.
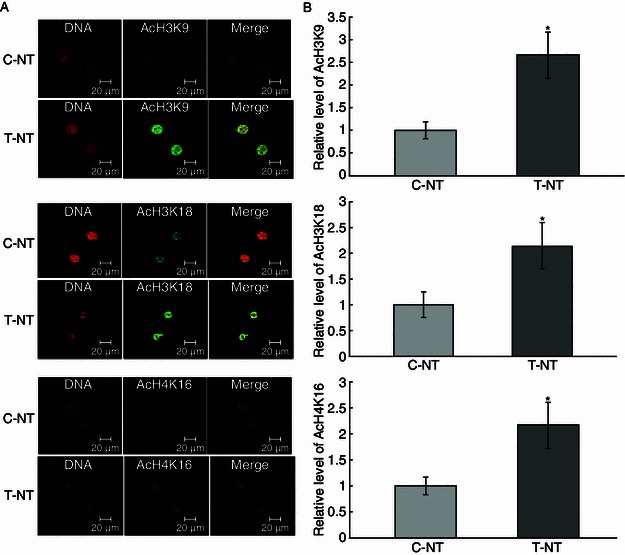
The global AcH3K9, AcH3K18, and AcH4K16 levels of 2-cell stage embryos. (A) Staining of AcH3K9, AcH3K18, and AcH4K16 (green) in non-treated control SCNT embryos (C-NT), and 2 μmol/L CBHA treated embryos (T-NT) at 2-cell stage. Each sample was counterstained with DAPI to visualize DNA (red). Original magnification was 200×. (B) Quantification of AcH3K9 and AcH3K18 signal intensities in C-NT (gray bars) and T-NT (black bars) embryos. Labeling intensity was expressed relative to that of the C-NT embryos (set as 100%). *P < 0.05, †P < 0.01. The experiments were replicated 3 times. n = 8–10 per group
Figure 4.
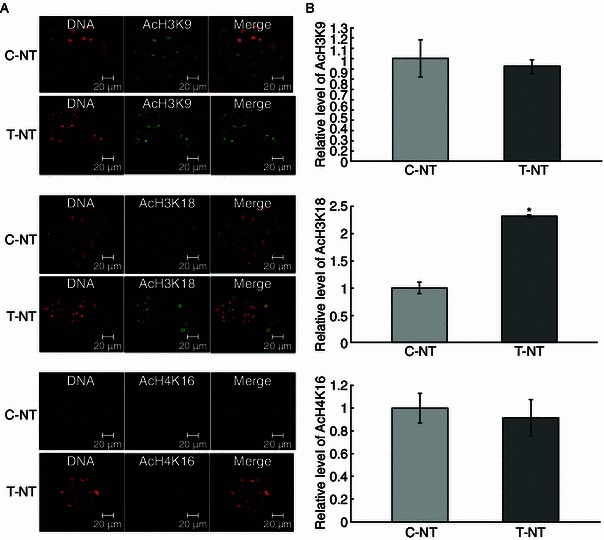
The global AcH3K9, AcH3K18, and AcH4K16 levels of blastocyst stage embryos. (A) Staining of AcH3K9, AcH3K18, and AcH4K16 (green) in non-treated control SCNT embryos (C-NT), and 2 μmol/L CBHA treated embryos (T-NT) at blastocyst stage. Each sample was counterstained with DAPI to visualize DNA (red). Original magnification was 200×. (B) Quantification of AcH3K9 and AcH3K18 signal intensities in C-NT (gray bars) and T-NT (black bars) embryos. Labeling intensity was expressed relative to that of the C-NT embryos (set as 100%). *P < 0.05. The experiments were replicated 3 times. n = 8–10 per group
Relative transcription level of development related genes, imprinted genes, and histone deacetylase genes
In mammalian early embryonic development, pou5f1 (oct4), an indicator for inner cell mass, is widely accepted to be a key factor in embryonic quality and pluripotency maintenance. Meanwhile, cdx2, a pivotal trophectoderm (TE) marker, is important for placenta formation. The expression level of imprinted genes and histone deacetylase genes is also vital for nuclear reprogramming. So, the transcription levels of the five genes (pou5f1, cdx2, igf2, igf2r, and hdac2) whose functions are mentioned above were detected by quantitive PCR (Fig. 5) at the blastocyst stage. The expression of two key developmental genes, pou5f1 and cdx2, was significantly increased by CBHA treatment. However, the expression of these two genes in both groups was significantly lower than the IVF (in vitro, fertilized) group. The expression of the imprinted gene IGF2 was increased after the CBHA treatment, which was similar to the IVF ones and significantly higher than the control group. However, the expression of another imprinted gene, igf2r, and the histone deacetylase gene, hdac2, was not affected by CBHA treatment, and both of them were significantly lower than the IVF ones.
Figure 5.
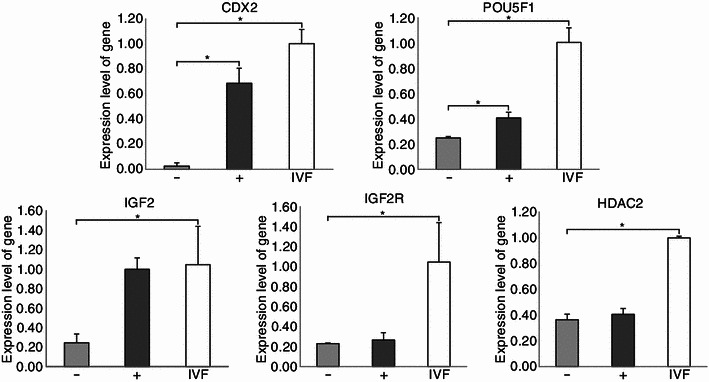
Relative expression at the transcriptional level of histone deacetylase genes, development-related, and imprinted genes. Blastocysts, IVF (open bars), non-treated control SCNT embryos (C-NT) (gray bars), and 2 μmol/L CBHA treated embryos (T-NT) (black bars) were collected on day 7. *P < 0.05; n = 15
Full term development of porcine SCNT embryos following CBHA treatment
To determine whether CBHA could improve the full term development of porcine SCNT embryos, 2-cell stage embryos with and without CBHA treatment were transferred to the oviducts of surrogates on the day of or 1 day after the onset of estrus. In the first batch of experiments, we transferred more than 200 embryos to each surrogate. We found that there was no significant difference between the two groups (Table 3), which is inconsistent with the beneficial effect of CBHA in the in vitro development. To further exclude the noise caused by the excessive number of embryos transferred, we designed a second batch of experiments, which transferred only 100 embryos to each recipient (Table 4). We obtained a total of 15 live, 6 dead, and 1 mummy from the CBHA treated group and 17 live, and 7 dead from the control group. The full term development ratio of CBHA-treated embryos was 4.2%, which is not significantly different to the control group (4.4%), suggesting CBHA treatment does not improve the full term development of porcine SCNT embryos.
Table 3.
Full-term development of CBHA treated SCNT embryos
| Cell type | No. of transferred embryos | CBHA treatmenta | Pregnancy checkb | Piglets born | Cloning efficiency (%)c |
|---|---|---|---|---|---|
| PEFCs | 231 | + | − | 0 | 0.94 |
| PEFCs | 187 | + | + | 2 dead + 1 mummy | |
| PEFCs | 276 | + | + | 1 live + 1 dead + 2 mummy | |
| PEFCs | 263 | + | + | 1 live + 1 dead | |
| PEFCs | 262 | − | − | 0 | |
| PEFCs | 280 | − | + | 2 live + 5 dead | 1.12 |
| PEFCs | 259 | − | + | 1 live + 3 dead | |
| PEFCs | 269 | − | + | 2 live + 1 dead | |
| PEFCs | 272 | − | + | 1 dead |
a+, Treated; −, untreated
b+, pregnant; −, not pregnant
cNo. of piglets/No. of embryos transferred
Table 4.
Full-term development of CBHA treated SCNT embryos with minimum transfer volume
| Cell type | No. of transferred embryos | CBHA treatmenta | Pregnancy checkb | Piglets born | Cloning efficiency (%)c |
|---|---|---|---|---|---|
| PEFCs | 100 | + | + | 6 live + 3 dead + 1 mummy | 4.2 |
| PEFCs | 100 | + | + | 6 live + 3 dead | |
| PEFCs | 100 | + | + | 3 live | |
| PEFCs | 100 | + | − | 0 | |
| PEFCs | 100 | + | − | 0 | |
| PEFCs | 100 | − | + | 5 live + 1 dead | 4.4 |
| PEFCs | 100 | − | + | 5 live + 1 dead | |
| PEFCs | 100 | − | + | 2 live | |
| PEFCs | 100 | − | + | 5 live + 3 dead | |
| PEFCs | 100 | − | − | 0 |
a+, Treated; −, untreated
b+, pregnant; −, not pregnant
cNo. of piglets/No. of embryos transferred
Discussion
Nowadays, somatic cells can be reprogrammed in many ways, the most common being somatic cell nuclear transfer (SCNT) and induced pluripotent stem cells (iPSCs) (Wilmut et al., 1997; Kato et al., 1998; Wakayama et al., 1998; Baguisi et al., 1999; Onishi et al., 2000; Chesne et al., 2002; Shin et al., 2002; Han et al., 2003; Galli et al., 2003; Woods et al., 2003; Zhou et al., 2003; Lee et al., 2005; Li et al, 2006; Takahashi et al., 2006; Yu et al., 2007). After being transfected with pluripotent genes or small molecules, a fraction of somatic cells could be reprogrammed to a pluripotent state like embryonic stem cells (ESCs). However, it has been reported that there are differences in the epigenetic modification and expression of genes between iPSCs and ESCs and the expression of exogenous pluripotent factors is likely to cause tumors (Okita et al., 2007; Bock et al., 2011; Menendez et al., 2011). Although several iPS cell lines have been reported, the silencing of exogenous genes and germline transmission remain unsolved. Thus nuclear transfer is still an indispensable method for somatic cell reprogramming. Meanwhile for pigs, besides therapeutic cloning, SCNT is a useful tool in transgenic research and especially in xenotransplantation and disease models. However, the efficiency of SCNT is still low and the mechanism of nuclear reprogramming is poorly understood.
To improve the efficiency of reprogramming, numerous factors or molecules have been used to alter the epigenetic modification in somatic cells or reconstructed embryos. HDACi is a widely used epigenetic modifier in cancer treatment and somatic cell nuclear reprogramming such as SCNT and iPSCs derivation. However, not all HDACi are beneficial to SCNT embryos, with some of them being unprofitable or detrimental. Generally, type II HDACi is used in epigenetic reprogramming (Monteriro et al., 2011).
Here we used CBHA, a hybrid polar compound, to improve the development of pig SCNT embryos. We found that the efficiency of SCNT and the quality of SCNT embryos are improved after CBHA treatment. Compared with non-treated embryos, CBHA treated SCNT embryos showed a higher global histone acetylation level. Expression of the development-related genes and the imprinted genes at blastocyst stage was also improved with the CBHA treatment.
In mouse SCNT, the optimum CBHA treatment protocol is 20 μmol/L incubation for 10 h post activation (Dai et al., 2010). However, in pig SCNT, we found that the best protocol was 2–10 μmol/L treated for 20–24 h after activation. This difference may be due to the differing developmental time spans between mice and pigs.
Histone acetylation is a significant epigenetic modification in embryo development and nuclear reprogramming. In previous studies, it has been shown that global hyperacetylation associated with a more permissive chromatin state which is an indicator of more fully reprogramming state in nuclear transfer and iPSCs (Li et al., 2002; Rybouchkin et al., 2006). In iPS cells, compared to partially reprogrammed ones, the fully reprogrammed iPSCs exhibit a more open and decondensed chromatin state and are more identical to ES cells (Mattout et al., 2011). We tested the histone acetylation level of reconstructed embryos after the CBHA treatment to examine the effect of this HDACi. Compared with the control SCNT embryos, the CBHA SCNT embryos showed a higher level of global histone acetylation. AcH3K9, AcH3K18, and AcH4K16 were all increased at the 1-cell and 2-cell stages, but there were no significant differences at the blastocyst stage except in AcH3K18. We speculated that CBHA depresses the expression of certain histone deacetylases, thus limiting the modification of deacetylation and increasing the global acetylation level. The hyperacetylation of histone may affect the chromatin remodeling and provide a more permissible and accessible configuration for transcription factors or chaperones combining (Vignon et al., 2002; Lee et al., 2004; Deshmukh et al., 2012; Mason et al., 2012). Embryonic gene activation (EGA) in pigs happened at the 4-cell stage (Maalouf et al., 2009), and the increase of global histone acetylation was mainly discovered at the 1-cell and 2-cell stages, which suggested that a more “open” chromatin structure might be beneficial for EGA.
To further explore the effect of CBHA on SCNT embryo development, we tested the expression of five important genes at the transcription level in the blastocysts of SCNT embryos, CBHA-SCNT embryos, and IVF embryos. This included two development-related genes: cdx2 and pou5f1, two imprinted genes: igf2 and igf2r, and histone deacetylase 2 (hdac2). In IVF embryos, the expression of cdx2 and pou5f1 are significantly higher than the control SCNT and CBHA treated SCNT embryos. However, CBHA treatment improved the expression of the two genes to a large extent compared with the control SCNT embryos. Cdx2 plays a key role in embryo placental development, and in mouse early embryo development, interaction between oct3/4 and cdx2 determines the level of trophectoderm differentiation (Tomanck et al., 1989; Chawengsaksophac et al., 2004; Strumpf et al., 2005). The extremely low expression of cdx2 in the control SCNT embryos may explain the abnormal placental development in some cloned embryos. While pou5f1 (oct4) is a well-known gene in embryo development, it interacts with regulatory enzymes and epigenetic histone markers in mice. As a central regulator of pluripotency and cell differentiation, it positively modulates the transcription of other recognized stem cell regulators, and it plays a vital role in inner cell mass (ICM) maintenance and ES cell self-renewal (Niwa et al., 2005; Loh et al., 2006). A previous study has shown that in porcine SCNT embryos, the expression of oct4 and oct4-related genes are aberrant (Pesce et al., 2001). It has been reported that in mice, CBHA could improve the derivation efficiency of ESCs from SCNT embryos (Dai et al., 2010). In this process, improved expression of pou5f1 may act. In our experiment, although still lower than the IVF embryos, the expression of pou5f1 in CBHA-SCNT embryos was significantly increased compared to the control SCNT embryos. For the importance of embryo development, the two best studied imprinted genes, igf2 and igf2r, have been discussed here. It has been reported that these two genes are related to EGA and involved in fetal growth regulation and are essential for normal development (Lee et al., 2006). Aberrant expression patterns of these imprinted genes may be responsible for the abnormal development found in fetuses and offspring originating from SCNT embryos (Latham et al., 1994; Yang et al., 2005). After CBHA treatment, we observed that the expression of igf2 increased to the same level as the IVF embryos, yet the expression of igf2r was not affected. It can be concluded that CBHA can improve the transcription activity of these genes, which should be related to the more “open” chromatin configuration. CBHA “corrects” the expression of the pivotal genes. It is known that histone deacetylase (hdacs) and histone acetylase (hats) mediate histone acetylation-deacetylation (Peserico et al., 2011). Previous studies on IVF and in vivo porcine embryos have reported that hdac2 exhibited a low level of expression at 4-cell stage, which steadily increased to its maximum level at the blastocyst stage, however this trend was reversed in SCNT embryos (Kumar et al., 2007; Peserico et al., 2011). Indeed, our results showed that the highest expression of hdac2 occurred in the IVF blastocyts. However, we also observed that the expression of hdac2 was hardly affected by the CBHA treatment. This may be because the hdac2 was not the target protein of CBHA and it is not the only deacetylase to regulate histone deacetylation.
Nevertheless, unlike the improved development in vitro, full term development efficiency was hardly affected by the CBHA treatment. The cloning efficiency between CBHA-SCNT and the control SCNT groups is not significantly different. One reason may be that the sample volume was not sufficient to show the difference between the two groups. Another may be because we transferred the embryos at the 2-cell stage. While the in vivo and in vitro conditions for pre-implanted embryo development are different, the in vivo environment may correct the aberrant development from the 2-cell stage onwards to some extent. Another possibility is that the donor cells we transferred are in a good state, therefore the embryos are well developed in vivo even without the CBHA treatment. Thus in our experiment, the piglet birth rate is much higher than other reports (0–3.7%) (Zhao et al., 2010), regardless of the CBHA-SCNT (4.2%) or the control SCNT groups (4.4%).
To sum up, our present study indicates that, as a HDACi, CBHA can improve the in vitro development of pig nuclear transfer embryos. The presence of CBHA can increase the global acetylation level in AcH3K9, AcH3K18, and AcH4K16. It influences the modification of chromatin structure and improves the expression of important genes in development and imprinting, rendering a more similar expression profile to IVF ones. It enhances the nuclear reprogramming and in vitro development of pig SCNT embryos, which may provide better qualified embryos for ES cell derivation in the case of low quality donors. Further studies are needed to discuss the effect of CBHA on chromatin configuration modifications.
Materials and methods
Chemicals
All chemicals were purchased from Sigma Chemical Co. (St. Louis, MO), unless otherwise stated.
Preparation of somatic cells for SCNT
Porcine fetal fibroblast cells were established from a 35-day old fetus. Cells at passage 2–5 were grown to 90% confluency in Dulbecco’s Modified Eagle Medium (DMEM; Gibco BRL, Life Technologies, Grand Island, USA) and supplemented with 10% fetal bovine serum (Gibco BRL, Paisley, UK). Fibroblast cells were dissociated with 0.25% trypsin-EDTA (Invitrogen, Carlsbad, CA, USA) at 37°C for about 3–5 min, then transferred to a centrifuge tube with Hepes-buffered Tissue Culture Medium 199 (TCM-199; BioWhittake, Walkersville, MD, USA) and supplemented with 2% cattle serum (CS; Danish Veterinary Institute, DTU, Frederiksberg, Denmark). The suspension was stored in ice until use.
In vitro maturation of oocytes
Ovaries were collected from prepubertal gilts at a local slaughterhouse and transported to the laboratory at 37°C. Follicles between 3 mm and 6 mm in diameter were aspirated with an 18-gauge needle attached to a 10-mL syringe. The cumulus-oocyte complexes (COCs) in the follicular fluid were allowed to settle by gravity. The COCs were rinsed in Hepes-buffered Tyrode’s medium containing 0.01% PVA three times. Only the COCs with multiple layers of intact cumulus cells and uniform ooplasm were selected for in vitro maturation (IVM). After washing three times in IVM medium, a group of 50 COCs was placed into each well of twenty-four-well cell culture plates containing 500 μL of IVM medium. The COCs were matured for 42 h at 38.5°C and 5% CO2 in air with 100% humidity. Matured COCs were then vortexed in 0.1% hyaluronidase in Hepes-buffered Tyrode’s medium containing 0.01% PVA for 3 min to remove the cumulus cells. Only the matured oocytes having an extruded first polar body (PB) with uniform cytoplasm were used for the SCNT or IVF embryos.
Somatic cell nuclear transfer
Matured oocytes used for SCNT were placed in manipulation medium (MAN) supplemented with 7.5 μg/mL Cytochalasin B, and overlaid with warm mineral oil. Oocytes were enucleated by aspirating the polar body (PB), metaphase II chromosomes, and a small amount of surrounding cytoplasm using a beveled glass pipette with an inner diameter of 17–20 μm. A single intact donor cell was injected into the perivitelline space and placed adjacent to the recipient cytoplasm. Karyoplast-cytoplast complexes (KCCs) were placed into embryo culture medium until fusion and activation occurred. The fusion and activation of the KCCs were accomplished with two direct current pulses (1 s interval) of 1.2 kV/cm for 30 microseconds provided by Eppendorf Multiporator in fusion medium (0.3 mol/L mannitol, 1.0 mmol/L CaCl2, 0.1 mmol/L MgCl2, and 0.5 mmol/L Hepes [pH adjusted to 7.0–7.4]). Oocytes were then incubated for 20 min in PZM3 and evaluated for fusion under a stereomicroscope. Only the fused embryos were placed into four-well cell culture plates containing 500 μL of PZM3 at 38.5°C and 5% CO2 with 100% humidified air.
Post-activation treatment and embryo culture
Stock solutions of CBHA were dissolved in dimethyl sulfoxide (DMSO) at 100 mmol/L and stored at −80°C. Following electrical activation, the SCNT embryos were treated with various concentrations (from 0.02 μmol/L to 400 μmol/L) of CBHA for different times (from 16 h to 24 h) in PZM3. After treatment, embryos were washed three times and then transferred into a four-well cell culture plate containing 500 μL PZM3 medium. They were cultured at 38°C and 5% CO2 with 100% humidity either overnight or for 6 days. Cleavage and blastocyst formation were evaluated on days 2 and 7, respectively, with the day of SCNT designated day 0.
Embryo transfer
Day 2 SCNT embryos were transferred to the oviducts of surrogates on the day of, or 1 day after, the onset of estrus. Pregnancy was diagnosed on day 25, and the surrogates were checked regularly at 2-week intervals by ultrasound examination. All of the cloned piglets were delivered by either natural birth or cesarean section on day 117 of gestation and hand raised. All animals were treated according to preapproved institutional animal care and use protocols.
Blastocyst cell number counting
Expanded day 6 blastocysts derived from SCNT were selected for cell number comparison. After being fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, embryos were mounted on slides in mounting medium containing 4,6-diamidino-2-phenylindole. At least 10 oocytes or embryos were processed for each separate sample, and the experiments were replicated three times. Slides were analyzed under an epifluorescent microscope (LEICA DM2500) equipped with a digital camera.
Detection of histone acetylation in SCNT embryos
Embryos treated without (control) or with the optimal CBHA concentration were collected at different stages of development: 1-cell stage (4 h after fusion); 2-cell stage (28–40 h after fusion); and blastocyst (140–150 h after fusion); and fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) for 1 h at 4–8°C. Subsequently, embryos were washed in PBS containing 0.1% Tween 20 (TPBS) for 30 min and permeabilized by 1% Triton X-100 in PBS overnight at 4–8°C. After washing, unspecific antibody binding was blocked by incubation in 2% bovine serum albumin (BSA) in PBS for 1 h, followed by 1 h incubation with primary antibody at RT (anti-acH3K9, 1:500; anti-acH4K16, 1:500; anti-acH3K18, 1:500). After washing, the embryos were incubated for 1 h with Alexa Flour 594-conjugated secondary antibody (1:100, Jackson, USA). Immunofluorescent labeling of AcH3K9, AcH3K18, and AcH4K16 in TSA treated and control embryos were evaluated under Zeiss LSM 510 Laser Scanning Microscope System with the same exposure times and adjustments of the microscope. Fluorescence was measured by analyzing the embryo pictures with Image J software. Average pixel intensity value was measured in 5 random nucleoplasm regions (excluding the nucleolar regions) subtracting the average value measured in the same process within the cytoplasm. Negative control was performed by omitting the primary antibody and led in all cases to the lack of labeling.
Quantitative PCR
At day 15, seven blastocysts were used per sample. The total RNA of the embryos was isolated using the Phenol-chloroform extraction protocol. The mRNA levels were quantified using SYBR Green Jump Start Taq Ready Mix on a real-time PCR detection system at the following thermal cycling conditions: 94°C for 2 min, followed by 40 PCR cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Transcripts were quantified in 3 replicates for each sample and calculated relative to the transcription of the housekeeping gene, β-actin, in every sample. The 2−△△CT was used to quantify the relative mRNA levels. The primers used in the quantitative PCR are listed in the Table S1.
Statistical analysis
Data were analyzed using SAS (9.1) with one-way ANOVA. A probability of P < 0.05 was considered to be statistically significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos.90919060 to Q.Z. and 31201112 to T.H.), and the “Strategic Priority Research Program” of the Chinese Academy of Sciences (Grant No. XDA01020101 to Q.Z.).
We thank Yuanxuan Zhao and Zechen Li for nuclear transfer and in vitro fertilization assistance; Xiangnan Miao for embryo transfer; and Shiwen Li for confocal microscopic studies.
Compliance with Ethics Guidelines
Yuran Song, Tang Hai, Ying Wang, Runfa Guo, Wei Li, Liu Wang, Qi Zhou declare that they have no conflict of interest.
All experiments comply with all of the relevant institutional and national animal welfare laws.
Abbreviations
- CBHA
m-carboxycinnamic acid bishydroxamide
- COCs
cumulus-oocyte complexes
- EGA
embryonic gene activation
- ESCs
embryonic stem cells
- HDACi
histone deacetylase inhibitors
- iPSC
induced pluripotent stem cells
- KCCs
karyoplast-cytoplast complexes
- PBS
phosphate buffered saline
- SCNT
somatic cell nuclear transfer
References
- Baguisi A, Behboodi E, Melican DT, Pollock JS, Destrempes MM, et al. Production of goats by somatic cell nuclear transfer. Nat Biotechnol. 1999;17:456–461. doi: 10.1038/8632. [DOI] [PubMed] [Google Scholar]
- Bendixen E, Danielsen M, Larsen K, Bendixen C, et al. Advances in porcine genomics and proteomics—a toolbox for developing the pig as a model organism for molecular biomedical research. Brief Funct Genomics. 2013;9(3):208–219. doi: 10.1093/bfgp/elq004. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawengsaksophac K, Graaff W, Beck F, et al. Cdx2 is essential for axial elongation in mouse development. PNAS. 2004;101:7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesne P, Adenot PG, Viglietta C, Baratte M, Boulanger L, et al. Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat Biotechnol. 2002;20:366–369. doi: 10.1038/nbt0402-366. [DOI] [PubMed] [Google Scholar]
- Dai XP, Hao J, Hou XJ, Hai T, Zhou Q, et al. Somatic nucleus reprogramming is significantly improved by m-carboxycinnamic acid bishydroxamide, a histone deacetylase inhibitor. J Biol Chem. 2010;285:31002–31010. doi: 10.1074/jbc.M110.136085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh RS, Ostrup O, Hyttel P, et al. Early aberrations in chromatin dynamics in embryos produced under in vitro conditions. Cell Reprogram. 2012;14:225–234. doi: 10.1089/cell.2011.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekser B, Ezzelarab M, Hara H, Windt D, Wijkstrom M, Bottino R, Trucco M, Cooper DKC, et al. Clinical xenotransplantation: the next medical revolution? Lancet. 2012;379:672–683. doi: 10.1016/S0140-6736(11)61091-X. [DOI] [PubMed] [Google Scholar]
- Galli C, Lagutina I, Crotti G, Colleoni S, Turini P, et al. A cloned horse born to its dam twin (vol 424, p 635, 2003) Nature. 2003;425:680. doi: 10.1038/424635a. [DOI] [PubMed] [Google Scholar]
- Gock H, Nottle M, Lew AM, d’Apice AJ, Cowan P et al (2011) Genetic modification of pigs for solid organ xenotransplantation. Transplant Rev 25:9–20 [DOI] [PubMed]
- Han YM, Kang YK, Koo DB, Lee KK, et al. Nuclear reprogramming of cloned embryos produced in vitro. Theriogenology. 2003;59:33–34. doi: 10.1016/S0093-691X(02)01271-2. [DOI] [PubMed] [Google Scholar]
- Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J, et al. Eight calves cloned from somatic cells of a single adult. Science. 1998;282:2095–2098. doi: 10.1126/science.282.5396.2095. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Ahn KS, Kim M, Shim H, et al. Comparison of potency between histone deacetylase inhibitors trichostatin A and valproic acid on enhancing in vitro development f porcine somatic cell nuclear transfer embryos. In Vitro Cell Dev Biol Anim. 2011;47:283–289. doi: 10.1007/s11626-011-9394-7. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Mizutani E, Ohta H, et al. Significant improvement of mouse cloning technique by treatment with trichostain A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Kumar BM, Jin HF, Kim JG, Ock SA, Hong Y, Balasubramanian S, et al. Differential gene expression patterns in porcine nuclear transfer embryos reconstructed with fetal fibroblasts and mesenchymal stem cells. Dev Dyn. 2007;236:435–446. doi: 10.1002/dvdy.21042. [DOI] [PubMed] [Google Scholar]
- Lager AE, Ragina NP, Cibelli JB, et al. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells. 2008;10:371–379. doi: 10.1089/clo.2007.0002. [DOI] [PubMed] [Google Scholar]
- Latham KE, Doherty AS, Scott CD, Schultz RM. Igf2r and Igf2 gene expression in androgenetic, gynogenetic, and parthenogenetic preimplantation mouse embryos: absence of regulation by genomic imprinting. Genes Dev. 1994;8:290–299. doi: 10.1101/gad.8.3.290. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hart SRL, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Lee BC, Kim MK, Jang G, Oh HJ, Yuda F, et al. Dogs cloned from adult somatic cells. Nature. 2005;436:641. doi: 10.1038/436641a. [DOI] [PubMed] [Google Scholar]
- Lee E, Lee SH, Lee BC, et al. Analysis of nuclear reprogramming in cloned miniature pig embryos by expression of Oct-4 and Oct-4 related genes. Biochem Biophys Res Commun. 2006;348:1419–1428. doi: 10.1016/j.bbrc.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Li E, et al. Chromatin modification and epigenetic reprogramming in mammalian development. Nature. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Li ZY, Sun XS, Chen J, Liu XM, Wisely SM, et al. Cloned ferrets produced by somatic cell nuclear transfer. Dev Biol. 2006;293:439–448. doi: 10.1016/j.ydbio.2006.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Svarcova O, Villemoes K, Vajta G, et al. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology. 2008;70:800–808. doi: 10.1016/j.theriogenology.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Ng HH, et al. The Oct4 and Nanog transcription network regulated pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Luo YL, Lin L, Bolund L, Jesen TG, Sorensen CB, et al. Genetically modified pigs for biomedical research. J Inherit Metab Dis. 2012;35:695–713. doi: 10.1007/s10545-012-9475-0. [DOI] [PubMed] [Google Scholar]
- Maalouf W, Liu ZC, Zink D, et al. Trichostatin A treatment of cloned mouse embryos improves constitutive heterochromatin remodeling as well as developmental potential to term. BMC Dev Biol. 2009;9:11. doi: 10.1186/1471-213X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason K, Liu ZC, Beaujean N, et al. Chromatin and epigenetic modifications during early mammalian development. Anim Reprod Sci. 2012;134:45–55. doi: 10.1016/j.anireprosci.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Mattout A, Biran A, Meshorer E, et al. Global epigenetic changes during somatic cell reprogramming to iPS cells. J Mol Biol. 2011;3:341–350. doi: 10.1093/jmcb/mjr028. [DOI] [PubMed] [Google Scholar]
- Menendez S, Camus S, Herreria A, et al. Increased dosage of tumor suppressors limits the tumorigenicity of cells without affecting their pluripotency. Aging Cell. 2011;11:41–50. doi: 10.1111/j.1474-9726.2011.00754.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Mori H, Mizobe Y, Sato M, et al. Benefical effects of reversine on in vitro development of miniature pig somatic cell nuclear transfer embryos. J Reprod Dev. 2010;56:291–296. doi: 10.1262/jrd.09-149A. [DOI] [PubMed] [Google Scholar]
- Monteriro FM, Oliveira CS, Oliveira LZ, Garcia JM et al (2011) Chromatin modifying agents in the vitro production of bovine embryos. SAGE-Hindawi access to research veterinary medicine international [DOI] [PMC free article] [PubMed]
- Niwa H, Toyooka Y, Rossant J, et al. Interaction between Oct3/4 and Cdx2 determines trophectodern differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Ogura A, Inoue K, Wakayama T, et al. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S, et al. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–318. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, et al. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000;289:1188–1190. doi: 10.1126/science.289.5482.1188. [DOI] [PubMed] [Google Scholar]
- Pesce M, Scholer HR, et al. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Peserico A, Simone C et al (2011) Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol 2011:371832 [DOI] [PMC free article] [PubMed]
- Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, Dal Y, Boone J, et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- Rybouchkin A, Kato Y, Tsunoda Y, et al. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol Reprod. 2006;74:1083–1089. doi: 10.1095/biolreprod.105.047456. [DOI] [PubMed] [Google Scholar]
- Shin T, Kraemer D, Pryor J, Liu L, Rugila J, et al. A cat cloned by nuclear transplantation. Nature. 2002;415:859. doi: 10.1038/nature723. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Rossant J, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, et al. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tomanck M, Kopecny V, Kanka J, et al. Genome reactivation in developing early embryos: an ultrastructural and autoradiaphic analysis. Anat Embryol. 1989;180:309–316. doi: 10.1007/BF00315889. [DOI] [PubMed] [Google Scholar]
- Verma N, Rettenmeier AW, Spanke SS, et al. Recent advances in the use of Sus scrofa (pig) as a model system for proteomic studies. Proteomics. 2011;11:776–793. doi: 10.1002/pmic.201000320. [DOI] [PubMed] [Google Scholar]
- Vignon X, Zhou Q, Renard JP, et al. Chromatin as a regulative architecture od the early developmental functions of mammalian embryos after fertilization or nuclear transfer. Cloning Stem Cells. 2002;4:363–377. doi: 10.1089/153623002321025041. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Perry ACF, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Walker SC, Shin T, Zaunbrecher GM, Romano JE, Johnson GA, Bazer FW, Piedrahita JA, et al. A highly efficient method for porcine cloning by nuclear transfer using in vitro-matured oocytes. Cloning Stem Cells. 2002;4:105–112. doi: 10.1089/153623002320253283. [DOI] [PubMed] [Google Scholar]
- Whitworth K, Prather RS, et al. Somatic cell nuclear transfer efficiency: how can it be improved through nuclear remodeling and reprogramming? Mol Reprod Dev. 2010;77:1001–1015. doi: 10.1002/mrd.21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William M, Rideout III, Eggan K, Jaenisch R, et al. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KHS. Viable offspring derived from fetal and adult mammalian cells (vol 385, p 810, 1997) Nature. 1997;386:200. doi: 10.1038/386200a0. [DOI] [PubMed] [Google Scholar]
- Woods GL, White KL, Vanderwall DK, Li GP, Aston KI, et al. A mule cloned from fetal cells by nuclear transfer. Science. 2003;301:1063. doi: 10.1126/science.1086743. [DOI] [PubMed] [Google Scholar]
- Yang L, Palmer PC, Tian XC, et al. Expression of imprinted genes is aberrant in deceased newborn cloned calves and relatively normal in surviving adult clones. Mol Reprod Dev. 2005;71:431–438. doi: 10.1002/mrd.20311. [DOI] [PubMed] [Google Scholar]
- Yu JY, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1916–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhao JG, Hao YH, Prather RS, et al. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell Reprogram. 2010;12:75–83. doi: 10.1089/cell.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Renard JP, Le Friec G, Brochard V, Beaujean N, et al. Generation of fertile cloned rats by regulating oocyte activation. Science. 2003;302:1179. doi: 10.1126/science.1088313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


