Abstract
The importance of commensal microbes for human health is increasingly recognized1-5, yet the impacts of evolutionary changes in human diet and culture on commensal microbiota remain almost unknown. Two of the greatest dietary shifts in human evolution involved the adoption of carbohydrate-rich Neolithic (farming) diets6,7 (beginning ~10,000 years BP6,8), and the more recent advent of industrially processed flour and sugar (~1850)9. Here, we show that calcified dental plaque (dental calculus) on ancient teeth preserves a detailed genetic record throughout this period. Data from 34 early European skeletons indicate that the transition from hunter-gatherer to farming shifted the oral microbial community to a disease-associated configuration. The composition of oral microbiota remained surprisingly constant between Neolithic and Medieval times, after which (the now ubiquitous) cariogenic bacteria became dominant, apparently during the Industrial Revolution. Modern oral microbiota are markedly less diverse than historic populations, which might be contributing to chronic oral (and other) disease in post-industrial lifestyles.
Commensal microbiota comprise the majority of cells in the body and play a key role in human health1-5,10. However, their evolution remains poorly understood, and detailed genetic records from commensal bacteria have yet to be recovered from the archaeological record. Dental calculus is ubiquitous in both present-day and ancient human populations11, and microscopic analysis has shown that it accurately preserves bacterial morphology over millennia12-14. Dental calculus develops when dental plaque, an extremely dense bacterial biofilm15, becomes mineralised with calcium phosphate16. Bacteria in calculus become locked in a crystalline matrix similar to bone16 (Supplementary Figure 1), with deposits occurring both above and below the gum or gingiva (supra- and subgingivally)17. Calculus represents one of the few sources of preserved human and hominid microbiota, and genetic analysis has the potential to create a powerful new record of past dietary impacts, health changes, and oral pathogen genomic evolution deep into the past. In addition, oral bacteria are transferred vertically from the primary caregiver(s) in early childhood18 and horizontally between family members later in life18,19, making archaeological dental calculus a potentially unique means of tracing population structure, movement and admixture between ancient cultures, as well as the spread of diseases.
The increased consumption of domesticated cereals (wheat and barley in the Near East) beginning with the Neolithic was associated with a marked increase in prevalence of dental calculus and oral pathology20. These oral diseases include dental caries (tooth decay)1 and periodontal disease (an infection causing damage to the supporting connective tissues of the tooth and resorption of bone)21, both of which were rare in pre-Neolithic hunter-gatherer societies20 and early hominins22. Caries and periodontal disease are both polymicrobial, plaque-mediated infections, thought to result from perturbation of a healthy, ecologically balanced oral biofilm23,24 that can occur from dietary changes, such as the increased consumption of fermentable carbohydrates25,26. Caries has become a major endemic disease, affecting 60-90% of school-aged children in industrialised countries, whilst periodontal disease occurs in 5-20% of the adult population worldwide27. Notably, oral bacteria are also associated with many systemic diseases including arthritis28, cardiovascular disease3 and diabetes4, in addition to diseases of the oral cavity1,2.
We collected a mixture of supra- and subgingival calculus samples (determined morphologically, Supplementary Note) from the teeth of 34 prehistoric European human skeletons (11 males, 11 females and 12 of unknown sex, ranging in age from < 20 to > 60 years at death, Supplementary Table 1), dating from before farming (Mesolithic) to Medieval periods. The samples included individuals from the last hunter-gatherers in Poland, the earliest farming culture in Central Europe (the Linear Pottery Culture – LBK), as well as late Neolithic (Bell Beaker Culture), early and later Bronze Age and medieval rural and urban populations (Table 1, see Supplementary Note for description). All ancient DNA work was conducted in a physically isolated, specialist laboratory dedicated to ancient environmental and bacterial DNA research at the Australian Centre for Ancient DNA, using strict decontamination and authentication protocols (Supplementary Note). We extracted bacterial DNA from sterilized ancient calculus samples (n = 34) and generated PCR amplicon libraries of the 16S rRNA gene, targeting three hypervariable regions (V1, V3 and V6) with barcoded primers (Supplementary Tables 2 and 3). In addition, primers specific to Streptococcus mutans and Porphyromonas gingivalis were used to detect oral pathogens in ancient dental calculus (Supplementary Table 2). We compared the ancient samples to modern calculus (n = 6) and plaque (n = 13) samples that were extracted and sequenced in an analogous manner (Methods, Supplementary Tables 3 and 4). We also extracted and sequenced bacterial DNA from within the teeth that provided the calculus samples, to determine the background bacterial contribution of the post-mortem depositional environment (n = 6, Supplementary Note). Amplicons generated from extracted samples and multiple extraction blanks were sequenced using both conventional and pyrosequencing technology. Of the 998,575 sequences generated, we discarded around ~ 50% following quality filtering and de-noising, to remove sequences containing PCR and sequencing errors29, leaving 451,241 sequences (Supplementary Table 4).
Table 1. Archaeological and anthropological samples used in the study.
| Sample ID | Museum# | Group/Culture | Period (years BP) | Location |
|---|---|---|---|---|
| 12011 | VI-1 B | Hunter-Gatherer | Mesolithic/para Neolithic (7550 - 5450) | Dudka, Poland |
| 12012 | VI-7 A | Hunter-Gatherer | Mesolithic/para Neolithic (7550 - 5450) | Dudka, Poland |
| 12013 | VI-14 A | Hunter-Gatherer | Mesolithic/para Neolithic (7550 - 5450) | Dudka, Poland |
| 12015 | VI-2 D | Hunter-Gatherer | Mesolithic/para Neolithic (7550 - 5450) | Dudka, Poland |
| 12016 | VI-1 A | Hunter-Gatherer | Mesolithic/para Neolithic (7550 - 5450) | Dudka, Poland |
| 12017 | VI-7 A | Hunter-Gatherer | Mesolithic/para Neolithic (7550 - 5450) | Dudka, Poland |
| 8215 | HK2000:4083a, 613.1 | LBK | Neolithic (7400 – 6725) | Halberstadt-Sonntagsfeld, Germany |
| 8240 | HK2000:4228a, 861 | LBK | Neolithic (7400 – 6725) | Halberstadt-Sonntagsfeld, Germany |
| 8247 | HK2000:4233a, 870 | LBK | Neolithic (7400 – 6725) | Halberstadt-Sonntagsfeld, Germany |
| 8275 | HK2000:7374a, 1324 | LBK | Neolithic (7400 – 6725) | Halberstadt-Sonntagsfeld, Germany |
| 8277 | HK2000:4014b, 413.1 | LBK | Neolithic (7400 – 6725) | Halberstadt-Sonntagsfeld, Germany |
| 4331 | HK2004:9463a, 6255.1 | Bell Beaker | Neolithic (4450 - 4000) | Quedlinburg XII, Germany |
| 9436 | HK, 43 | LN/BA | LN/BA (4150 – 3600) | Benzingerode-Heimburg, Germany |
| 8890 | T82GF | Bronze Age | Bronze Age | Yorkshire, England |
| 8891 | 14Barrow 163 | Bronze Age | Bronze Age | Yorkshire, England |
| 8894 | T98 | Bronze Age | Bronze Age | Yorkshire, England |
| 8326 | 2095 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8330 | 2106 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8332 | 4440 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8477 | 2357 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8482 | 2654 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8814 | 4161 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8863 | 4485 | Jewbury | Late Medieval (750 - 650) | York, England |
| 8333 | R5287 | Raunds Furnells | Early Medieval (1100 – 850) | Northamptonshire, England |
| 8335 | R5252 | Raunds Furnells | Early Medieval (1100 – 850) | Northamptonshire, England |
| 8337 | R5136 | Raunds Furnells | Early Medieval (1100 – 850) | Northamptonshire, England |
| 8341 | R5206 | Raunds Furnells | Early Medieval (1100 – 850) | Northamptonshire, England |
| 8868 | R5157 | Raunds Furnells | Early Medieval (1100 – 850) | Northamptonshire, England |
| 8869 | R5229 | Raunds Furnells | Early Medieval (1100 – 850) | Northamptonshire, England |
| 8873 | 5228 | StHW | Late Medieval (1000 - 400) | York, England |
| 8874 | 5241 | StHW | Late Medieval (1000 - 400) | York, England |
| 8877 | 5113 | StHW | Late Medieval (1000 - 400) | York, England |
| 8878 | 5203 | StHW | Late Medieval (1000 - 400) | York, England |
| 8883 | 5244 | StHW | Late Medieval (1000 - 400) | York, England |
| 1 | NA | European Descent | 0 | Adelaide, Australia |
| 2 | NA | European Descent | 0 | Adelaide, Australia |
| 3 | NA | European Descent | 0 | Adelaide, Australia |
| 4 | NA | European Descent | 0 | Adelaide, Australia |
| 5 | NA | European Descent | 0 | Adelaide, Australia |
| 6 | NA | European Descent | 0 | Adelaide, Australia |
| 7 | NA | European Descent | 0 | Adelaide, Australia |
| 8 | NA | European Descent | 0 | Adelaide, Australia |
| 9 | NA | European Descent | 0 | Adelaide, Australia |
| 10 | NA | European Descent | 0 | Adelaide, Australia |
Abbreviations: LBK - Linear Pottery Culture; LN/BA - Late Neolithic/early Bronze Age; StHW - St. Helen-on-the-Walls. Mesolithic/para-Neolithic is the terminology used for the transitional cultures of the forest zone of Eastern Europe. See Supplementary Table 1 for further information about the ancient calculus, and modern plaque and calculus samples.
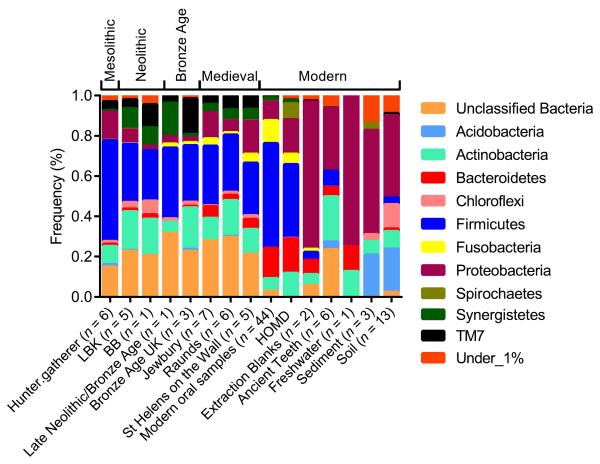
At the phylum level, the bacterial composition of ancient calculus was similar to modern oral samples and sequences from the Human Oral Microbiome Database (HOMD)30, but markedly distinct from laboratory reagents (extraction blanks), environmental samples (soils, sediments and water), and within the ancient teeth themselves (Figure 1, Supplementary Fig. 2 and Supplementary Note). The archaeological calculus was dominated by Firmicutes (33% for the V3 region, which was the most phylogenetically informative fragment, see Supplementary Note), which is comparable to percentages found in both the HOMD (37%), and modern oral samples (average 50%)1,2,21,31. Again, this was clearly distinguishable from the bacterial sequences obtained from extraction blanks (6% Firmicutes, p = 0.003), environmental samples (1.6% Firmicutes, p < 0.001) and within the ancient teeth (8% Firmicutes, p < 0.001), which were all dominated by Proteobacteria (73% p = 0.2, 56% p < 0.001 and 31% p = 0.005, respectively). The sequences from the extraction blanks were typical of bacterial communities found in clean-room environments32 and non-template controls33. In addition to Firmicutes, the ancient dental calculus samples contained all 15 phyla commonly found in the modern human oral cavity30, with high percentages of Actinobacteria (19%) as is observed in modern calculus deposits (7%). We have shown that dental calculus from samples that are thousands of years old preserve representative and informative microbial signatures of past human-associated microbiota.
Figure 1. Phylum level microbial composition of the ancient dental calculus deposits.
This is similar to modern oral samples, and distinct from non-template controls (EBC), ancient human teeth and environmental samples. The phylum frequencies for the V3 region are presented for the ancient calculus samples (LBK; Linear Pottery Culture, BB; Bell Beaker), modern oral samples, which included pyrosequenced (calculus, plaque and saliva31) and cloned (plaque1,2,21) data, non-template controls (or extraction blanks), ancient human teeth and environmental samples (freshwater, sediments and soils34-40) (Supplementary Table 1). Phylum frequencies from the HOMD were generated from partial and full-length sequences of the 16S rRNA gene. The phyla with a frequency < 1% include: ABY1_OD1, AD3, Armatimonadetes, BRC1, CCM11b, Chlamydiae, Chlorobi, Cyanobacteria, Elusimicrobia, Euryarchaeota, Fibrobacteres, GAL15, Gemmatimonadetes, GN02, GN04, GOUTA4, KSB1, Lentisphaerae, NC10, Nitrospirae, NKB19, OP11, OP3, OP9, PAUC34f, Planctomycetes, SBR1093, SC3, SC4, SM2F11, SPAM, Spirochaetes, SR1, Tenericutes, Thermi, TM6, Verrucomicrobia, WPS-2, WS3 and ZB2.
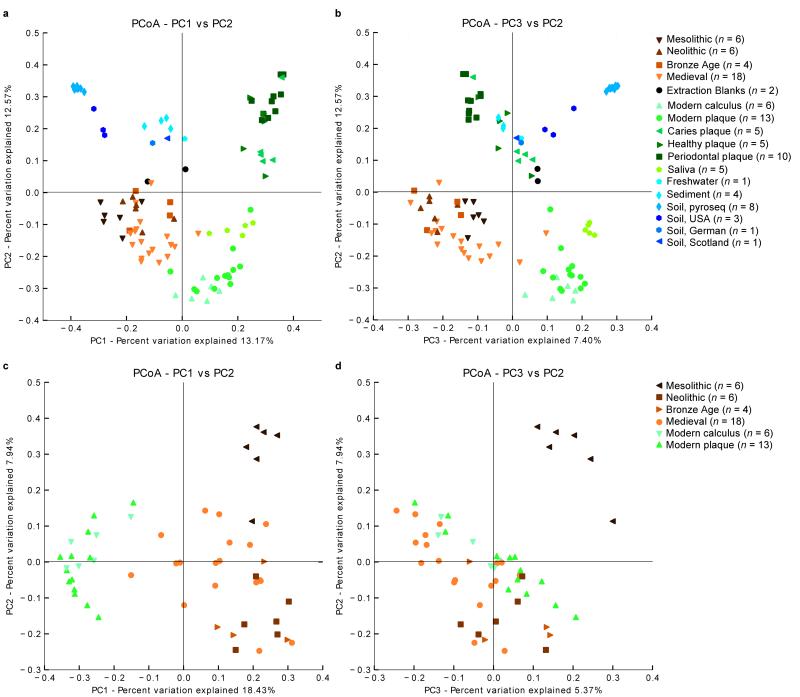
Phylogenetic analyses of the β-diversity, which measures the number of operational taxonomic units (OTUs) that are unique between the groups (Supplementary Note), confirmed the V3 sequences from ancient calculus were clearly more similar to those from modern dental calculus, plaque, and saliva samples1,2,21,31 than environmental samples34-40 (Figure 2a and b, Supplementary Fig. 4 and Supplementary Table 5). Similar patterns were observed for sequences from V1 and V6 (Supplementary Fig. 5, Supplementary Note). Furthermore, the bacterial composition of the ancient calculus samples clustered separately from the sequences present within the ancient teeth (Supplementary Fig. 6, p = 0.002). Overall, these results strongly suggest that the DNA sequences from the ancient calculus samples are not derived from contamination in the post-mortem environment.
Figure 2. Principal components plot of β-diversity.
PCoA reveals a close phylogenetic relationship between ancient dental calculus and modern oral samples, both of which are distinct from the non-template controls and environmental samples. β-diversity was calculated for all samples (Supplementary Note) using the UniFrac metric for the V3 region, and PCoA was applied to the unweighted, UniFrac distances. The plot of the first and second components (PC1 and PC2) (a) and the first and third components (PC1 and PC3) (b) of the PCoA clustered the ancient dental calculus samples with the modern oral pyrosequenced data (calculus, plaque and saliva), which were separated from the environmental samples and extraction blanks. A restricted PCoA plot of PC1 and PC2 (c) and PC1 and PC3 (d) that only includes ancient and modern oral pyrosequencing samples separated the hunter-gatherer (Mesolithic) individuals from modern, Medieval, and Neolithic samples.
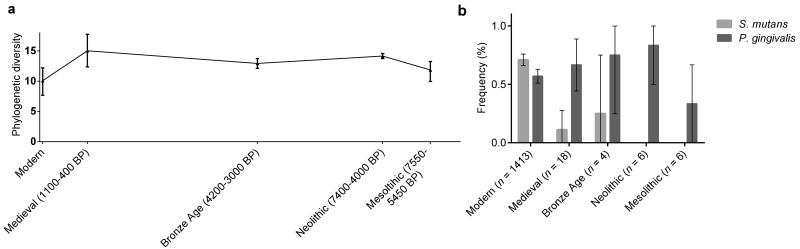
The temporal transect of ancient dental calculus samples provides the first views about the timing and nature of changes in human oral bacterial composition and diversity over the last 7,500 years. The composition of oral microbiota underwent a distinct shift with the introduction of farming in the early Neolithic (Figures 2c and d), with the earlier hunter-gatherer groups displaying fewer caries- and periodontal disease-associated taxa (Figure 3). This is consistent with skeletal evidence showing marked increases in periodontal disease41 following the transition to an agricultural diet, suggesting a major impact on the human oral ecosystem around this time. This is thought to be caused by increased amounts of soft carbohydrate foods compared with hunter-gatherer diets26. After the transition to agriculture in the early Neolithic there was a notable consistency in the composition of bacteria through the Medieval period (~ 400 years BP) (Figure 3), in parallel with the broad similarity of food-processing technologies during these times9. In contrast, today’s oral environment is much less biodiverse and dominated by potentially cariogenic bacteria (Figure 3a and b, e.g., S. mutans, Supplementary Figs. 7-9 and Supplementary Table 7).
Figure 3. Changes in the diversity and composition of oral microbiota.
(a) For the V3 region sequences, we estimated the phylogenetic diversity50 (Supplementary Note) of the archaeological dental calculus samples (n = 34) and compared them to modern calculus (n = 6) and plaque (n = 13). We estimated phylogenetic diversity from only classified, Gram-positive bacterial sequences to minimize the influence of taphonomic bias (see Supplementary Note). Diversity was calculated at a depth of 34 sequences and bootstrapped to assess the robustness of the pattern. Error bars represent bootstrapped frequencies generated by sampling 255 replicates without replacement. (b) Specific primers were used to amplify sequences unique to the oral pathogens S. mutans (light) and P. gingivalis (dark).
Random forest (RF) analysis was used to identify the taxa which discriminate the different time periods (Supplementary Note). RF revealed that Clostridia taxa, such as Clostridiales (importance score = 0.014 ± 0.002) and non-pathogenic oral microbial family Ruminococcaceae (importance score = 0.0035 ± 0.0009), were predictive of hunter-gatherer microbial communities compared to early agriculturists (ratio baseline error to observed error = 3.5, Supplementary Table 8). Farming groups from the Neolithic and Medieval periods were discriminated by both non-pathogenic taxa such as Clostridiales incertae sedis (importance score = 0.014 ± 0.003) and decay-associated Veillonellaceae (importance score= 0.012 ± 0.0038). Farming populations also displayed more periodontal disease-associated taxa, including P. gingivalis, Tannerella and Treponema, than hunter-gatherers. While there is also a strong association between periodontal disease and individual age at death42, we found periodontal disease-associated taxa across a range of ages, including the youngest individual (3-4 years old, ID 8247).
RF analysis revealed that only a limited number of taxa distinguished modern oral environments from farming groups in the Medieval and Neolithic periods (ratio baseline error to observed error = 4.0 , Supplementary Table 9). These taxa include decay-associated Veillonellaceae (importance score = 0.021 ± 0.004), in addition to Lachnospiraceae (importance score= 0.019 ± 0.007) and Actinomycetales (importance score = 0.0013 ± 0.0005). Notably, the frequency of S. mutans is significantly higher in modern samples than pre-industrial, agricultural samples (Figure 3b, Supplementary Note), indicating that caries-associated bacteria have only become dominant after Medieval times. This change is most likely associated with the onset of the Industrial Revolution which began some 200 years ago and represents the largest change in food production and processing technology since the shift to farming9. The Industrial Revolution saw the production of refined grain and concentrated sugar from processed sugar beet and cane9, generating mono- and disaccharides, which are the main substrates for microbial fermentation that lowers plaque pH and cause enamel demineralization26.
Overall, it is clear that modern Europeans have much lower oral microbial diversity than either Mesolithic or pre-Industrial Neolithic groups (p < 0.001, Supplementary Table 7), including fewer bacteria associated with good health (Ruminococcaceae), periodontal disease-associated taxa (e.g., Porphyromonas gingivalis, Tannerella and Treponema) similar to early agriculturists, and a markedly higher abundance of (now ubiquitous) pathogens such as Streptococcus mutans (Figure 2, Supplementary Note). Perhaps more importantly, the decline in overall oral microbial diversity indicates that over the past few hundred years, the human mouth has become a substantially less-biodiverse ecosystem. In both a human-associated microbiota43-45 and macroecological46,47 context, higher phylogenetic diversity is associated with greater ecosystem resilience and productivity. Therefore, the modern oral environment is likely to be less resilient to perturbations48 in the form of dietary imbalances or invasion49 by pathogenic bacterial species.
Major changes in carbohydrate intake in human history appear to have impacted the ecosystem of the mouth, opening up pathological niches for periodontal disease in the early Neolithic and caries in the recent past. These data are potentially important for assessing current associations with systemic diseases; for example, it has been proposed that periodontal disease might contribute to the development of diabetes and heart disease25 through the production of a prolonged inflammatory state3. However, whilst the frequency of these systemic diseases has risen over the last few decades25, our data show that the abundance of periodontal disease-associated bacteria has been relatively stable since the introduction of farming (e.g., Figure 3, P. gingivalis). This indicates that although periodontal disease might contribute to pathogenesis, it is probably not a factor in the rising incidence of these systemic diseases.
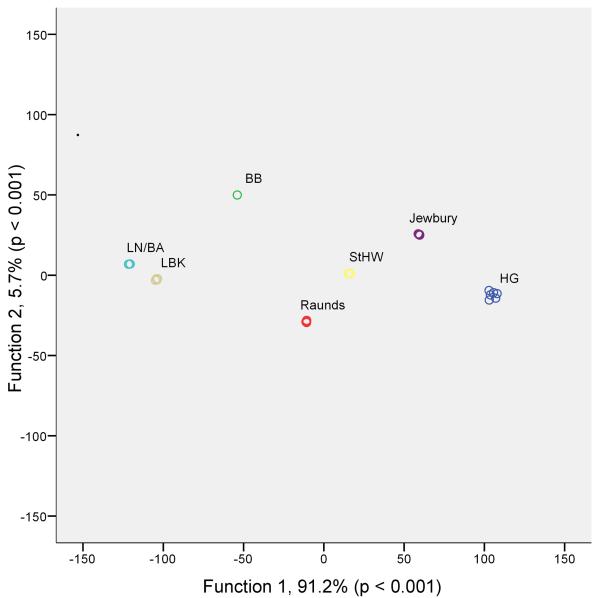
Our research has identified a powerful new avenue for bio-anthropological research, which promises to provide the first detailed genetic records of the evolution of human microbiota. This provides the potential to directly examine the effects of nutritional and cultural transitions (see Figure 4, Supplementary Figs. 10 and 11 and Supplementary Note) on human health through time and to record the genomic evolution of human commensals and pathogens.
Figure 4. Discriminant analysis of β-diversity.
Discriminant analysis was applied to the principal coordinates generated from the unweighted UniFrac distances calculated from the V3 region sequences. Each individual is represented by a circle and coloured according to archaeological grouping (HG; Hunter-Gatherer, LBK; Linear Pottery Culture, BB; Bell Beaker, LN/BA; Late Neolithic/Bronze Age, StHW; St. Helen-on-the-Walls). The majority of phylogenetic variation (91.2%) was described by the first discriminant function, showing that individuals from the same archaeological groups cluster according to microbial composition
Methods
The ancient dental calculus sample details, including archaeological information, preparation methodology and authentication criteria, are described in Supplementary Note.
DNA extraction, PCR, cloning and sequencing
We extracted DNA from 0.05–0.2 g of sterilized and powdered ancient dental calculus, and included a non-template control every three extractions. The ancient dental calculus deposits, modern calculus and plaque samples, and non-template controls were lysed in 1 ml of lysis buffer, containing 0.5 M EDTA (pH 8), SDS (10%) and Proteinase K (20 mg/ml). The samples and lysis buffer were rotated for 24 hours at 55 °C. Following sample lysis, DNA was isolated using the QIAamp DNA Investigator Kit (Qiagen). DNA was eluted to a final volume of 100 μl, and extracts were stored at 4 °C. The tooth samples were extracted using protocols described previously51. Independent extractions were not possible due to the small size of samples; commonly, only one calculus deposit per individual was available for DNA analysis.
PCR was used to amplify microbial DNA in the ancient dental calculus samples, modern oral samples, tooth samples and extraction controls using both universal 16S rRNA gene microbial primers and specific primers for the oral pathogens, Streptococcus mutans (GtfB gene) and Porphyromonas gingivalis (16S rRNA gene) (Supplementary Table 2). We also attempted (unsuccessfully) to amplify human mtDNA from the ancient dental calculus samples. For all primer sets, the PCR conditions were as follows; Amplitaq Gold (Applied Biosystems) at 2 U in 25 μl volumes using 1× Buffer Gold, 2.5 mM MgCl2, 0.25 mM of each dNTP (Fermentas), 400 μM of each primer, 1 mg/ml RSA (Sigma-Aldrich), ShrimpDNAse (Affymetrix) at 0.002 U/μl and 2 μl of DNA extract. ShrimpDNase was used to remove microbial contamination from PCR reagents prior to the amplification reaction, and was added to the PCR mixture (minus the extract), incubated at 37 °C for 15 minutes, and then inactivated by heating the mixture to 65 °C for 15 minutes. For the specific primers, the thermocycling conditions consisted of an initial enzyme activation at 95 °C for 6 minutes, followed by 45 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s and elongation at 72 °C for 30 s and a single final extension step at 60 °C for 10 minutes. Only 40 cycles were used to amplify 16S rRNA universal sequence, and the annealing temperature was 50 °C. Each set of PCRs included multiple extraction and PCR blanks. All PCRs were visually examined by electrophoresis on 3.5% agarose TBE gels. Specific PCR products were purified using 5 μl of amplified product, exonuclease I (0.8 U/μl) and shrimp alkaline phosphatase (1 U/μl). The mixture was heated to 37 °C for 40 minutes and then heat inactivated at 80 °C for 10 minutes. The purified amplicons were sequenced bidirectionally using PCR primers and the BigDye Terminator 3.1 Kit (Applied Biosystems), according to the manufacturer’s instructions. Sequencing products were purified using a MultiscreenHTS Vacuum Manifold (Millipore), according to the manufacturer’s protocol. Sequencing products were separated on the 3130×l Genetic Analyzer (Applied Biosystems), and the resulting sequences were edited using Sequencher (version 4.7).
We cloned the 16S rRNA gene universal amplicons to monitor contamination within the ancient samples and non-template controls and to assess the suitability of calculus samples for 454 sequencing. The PCR products were purified using Agencourt Ampure (Beckman Coulter) according to the manufacturer’s instructions, and cloned using a StrataClone PCR cloning Kit (Stratagene). The clones were added directly to the colony PCR mix, which contained 10× Hotmaster Buffer (Eppendorf), Hotmaster Taq 0.5 U/μl (5Prime), forward and reverse M13 primers (10 μM) (Supplementary Table 2) in a 25 μl reaction. The thermocycling conditions consisted of 94 °C for 10 minutes and 35 cycles of 94 °C for 20 s, 55 °C for 10 s and 65 °C for 45 s, with a single extension of 65 °C for 10 minutes. Colony PCRs were visually inspected on 2% agarose TBE gels. The colony PCR products were purified and sequenced using the same protocols as described for the products of the specific primers.
454 GS FLX Titanium Sequencing
Pyrosequencing (GS-FLX Titanium) was used to examine 34 ancient dental calculus samples, 19 modern oral samples (six calculus and 13 plaque), six tooth samples and two extraction blanks. For the ancient calculus, modern oral samples and non-template controls, three hypervariable regions of the 16S rRNA gene (V1, V3 and V6) were amplified, using the conditions described above. For the tooth samples, only the V3 region was amplified, again using the same conditions. The forward and reverse primers contained 454 Lib-L kit A- and B-adaptors, respectively, at the 5′ end. The forward primer also contained sample-specific barcodes (Supplementary Table 3) that were developed by the Human Microbiome Project (www.hmpdacc.org/tools_protocols/tools_protocols.php). The barcode sequences had not previously been used in either the Australian Centre for Ancient DNA or the Wellcome Trust Sanger Institute, where the 454 sequencing was performed. Hence, all sequences retrieved that did not contain a barcode were assumed to be contaminants and discarded. Each region of the 16S rRNA gene was amplified twice (on different days), and the duplicates were pooled for 454 sequencing to minimize the potential impact of preferential sequence amplification.
Filtering, OTU picking, alignment and taxonomic assignment of 454 sequences
The sequences from the GS FLX Titanium platform were processed using the QIIME (version 1.5.0) software package52. Quality filtering was performed to remove sequences which were either under 60 bp (potential primer dimers), contained ambiguous bases, had primer or barcode mismatches, contained homopolymers which exceeded six bases or had an average quality score below 25. The remaining sequences ranged between 60 and 210 bp in length. The quality-filtered sequences were denoised53 to remove sequences containing errors produced during the PCR (single base errors) and pyrosequencing, which resulted in the removal of ~ 50% of the sequences that were identified as having ambiguous flow data (Supplementary Table 4). However, we found that sequence classifications and beta diversity analyses were comparable between the dataset on which only quality-filtering had been performed and the de-noised dataset, as has previously been shown53. Similar sequences were binned into OTUs using optimal UCLUST54 at a 95% likeness. Clustering is more commonly performed at 97%; however, a 95% cut-off has been found to classify OTUs more accurately for closely related, short sequences55. Representative sequences from each OTU were aligned using PyNAST52 against the GreenGenes core set, with a minimum length of 60 bp and identity of 75%. PyNAST aligns the short GS FLX generated sequences (60-210 bp) against the full 16S rRNA gene. Columns which solely contained gaps were removed from the alignment prior to building phylogenetic trees. To overcome the difficulty in aligning highly variable 16S rRNA gene sequences, it is common to hide or lane-mask regions where at least 50% of the base composition is not conserved56. We did not hide variable regions because lane-masked alignments can ‘mute’ the phylogenetic diversity observed55. The gap-filtered sequences were taxonomically assigned using the RDP classifier and nomenclature57.
Detailed descriptions of the analyses performed on the ancient dental calculus, modern oral and extraction blank sequences are described in the Supplementary Note: β-diversity, alpha-diversity, Random Forest and discriminant analyses.
Supplementary Material
Acknowledgements
We thank N. Gully and S. Bent for critical discussions, J. Soubrier for bioinformatics assistance, the Australian Research Council, Wellcome Trust (WT092799MA and WT076964) and the Sir Mark Mitchell Foundation for funding support. We thank Harald Meller from the State Heritage museum of Saxony-Anhalt, Germany for prehistoric samples and members of the Australian Centre for Ancient DNA for practical help, and providing samples of plaque and calculus. We thank several anonymous reviewers whose comments have considerably improved the manuscript.
Footnotes
Sequence data have been deposited in Genbank under accession ERP002107.
The authors declare no competing financial interests.
References
- 1.Aas JA, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–17. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dave S, Van Dyke T. The link between periodontal disease and cardiovascular disease is probably inflammation. Oral Dis. 2008;14:95–101. doi: 10.1111/j.1601-0825.2007.01438.x. [DOI] [PubMed] [Google Scholar]
- 4.Grossi SG, Genco RJ. Periodontal disease and diabetes mellitus: a two-way relationship. Ann Periodontol. 1998;3:51–61. doi: 10.1902/annals.1998.3.1.51. [DOI] [PubMed] [Google Scholar]
- 5.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braidwood RJ, Howe B, Reed CA. The Iranian Prehistoric Project: New problems arise as more is learned of the first attempts at food production and settled village life. Science. 1961;133:2008–10. doi: 10.1126/science.133.3469.2008. [DOI] [PubMed] [Google Scholar]
- 7.Oelzea VM, et al. Early Neolithic diet and animal husbandry: stable isotope evidence from three Linearbandkeramik (LBK) sites in Central Germany. Journal of Archaeological Science. 2011;38:270–279. [Google Scholar]
- 8.Childe VG. The dawn of European civilisation. Kegan Paul; London: 1925. [Google Scholar]
- 9.Cordain L, et al. Origins and evolution of the Western diet: health implications for the 21st century. Am J Clin Nutr. 2005;81:341–54. doi: 10.1093/ajcn.81.2.341. [DOI] [PubMed] [Google Scholar]
- 10.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 11.Scott GR, Poulson SR. Stable carbon and nitrogen isotopes of human dental calculus: a potentially new non-destructive proxy for paleodietary analysis. Journal of Archaeological Science. 2012;39:1388–1393. [Google Scholar]
- 12.Lilley J, Stroud G, Brothwell D. The Jewish burial ground at Jewbury. In: Addyman PV, Kinsler VA, editors. The Arcaheology of York. Vol. 12. Council for British Archaeology; York: 1994. [Google Scholar]
- 13.Preus HR, Marvik OJ, Selvig KA, Bennike P. Ancient bacterial DNA (aDNA) in dental calculus from archaeological human remains. Journal of Archaeological Science. 2011 [Google Scholar]
- 14.Vandermeersch B, et al. Middle Palaeolithic dental Bacteria from Kebara, Israel. C.R. Acad Sci. Paris. 1994;319:727–731. [Google Scholar]
- 15.Socransky SS, Haffajee AD. Dental biofilms: difficult therapeutic targets. Periodontol 2000. 2002;28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y, Yip HK. Supragingival calculus: formation and control. Crit Rev Oral Biol Med. 2002;13:426–41. doi: 10.1177/154411130201300506. [DOI] [PubMed] [Google Scholar]
- 17.Lieverse AR. Diet and the aetiology of dental calculus. International Journal of Osteoarchaeology. 1999;9:219–232. [Google Scholar]
- 18.Asikainen S, Chen C, Slots J. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol Immunol. 1996;11:387–94. doi: 10.1111/j.1399-302x.1996.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Steenbergen TJ, Menard C, Tijhof CJ, Mouton C, De Graaff J. Comparison of three molecular typing methods in studies of transmission of Porphyromonas gingivalis. J Med Microbiol. 1993;39:416–21. doi: 10.1099/00222615-39-6-416. [DOI] [PubMed] [Google Scholar]
- 20.Aufderheide AC, Rodriguez-Martin C, Langsjoen O. The Cambridge encyclopedia of human paleopathology. Cambridge University Press; Cambridge: 1998. [Google Scholar]
- 21.Faveri M, et al. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. 2008;23:112–8. doi: 10.1111/j.1399-302X.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- 22.Grine FE, Gwinnett AJ, Oaks JH. Early hominid dental pathology: interproximal caries in 1.5 million-year-old Paranthropus robustus from Swartkrans. Arch Oral Biol. 1990;35:381–6. doi: 10.1016/0003-9969(90)90185-d. [DOI] [PubMed] [Google Scholar]
- 23.Marsh PD. Sugar, fluoride, pH and microbial homeostasis in dental plaque. Proc Finn Dent Soc. 1991;87:515–25. [PubMed] [Google Scholar]
- 24.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–94. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 25.Hujoel P. Dietary carbohydrates and dental-systemic diseases. J Dent Res. 2009;88:490–502. doi: 10.1177/0022034509337700. [DOI] [PubMed] [Google Scholar]
- 26.Marsh PD. Microbiology of dental plaque biofilms and their role in oral health and caries. Dent Clin North Am. 2010;54:441–54. doi: 10.1016/j.cden.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C. The global burden of oral diseases and risks to oral health. Bull World Health Organ. 2005;83:661–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Mercado FB, Marshall RI, Klestov AC, Bartold PM. Relationship between rheumatoid arthritis and periodontitis. J Periodontol. 2001;72:779–87. doi: 10.1902/jop.2001.72.6.779. [DOI] [PubMed] [Google Scholar]
- 29.Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–41. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 30.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics. 2010;11:523. doi: 10.1186/1471-2164-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Duc MT, Kern R, Venkateswaran K. Microbial monitoring of spacecraft and associated environments. Microb Ecol. 2004;47:150–8. doi: 10.1007/s00248-003-1012-0. [DOI] [PubMed] [Google Scholar]
- 33.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 34.Beier S, Witzel KP, Marxsen J. Bacterial community composition in Central European running waters examined by temperature gradient gel electrophoresis and sequence analysis of 16S rRNA genes. Appl Environ Microbiol. 2008;74:188–99. doi: 10.1128/AEM.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schloss PD, Handelsman J. Toward a census of bacteria in soil. PLoS Comput Biol. 2006;2:e92. doi: 10.1371/journal.pcbi.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis RJ, Morgan P, Weightman AJ, Fry JC. Cultivation-dependent and - independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol. 2003;69:3223–30. doi: 10.1128/AEM.69.6.3223-3230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nogales B, et al. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl Environ Microbiol. 2001;67:1874–84. doi: 10.1128/AEM.67.4.1874-1884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elshahed MS, et al. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol. 2008;74:5422–8. doi: 10.1128/AEM.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 40.Will C, et al. Horizon-Specific Bacterial Community Composition of German Grassland Soils, as Revealed by Pyrosequencing-Based Analysis of 16S rRNA Genes. Applied and Environmental Microbiology. 2010;76:6751–6759. doi: 10.1128/AEM.01063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerr NW. Prevalence and natural history of periodontal disease in prehistoric Scots (pre-900 AD) J Periodontal Res. 1998;33:131–7. doi: 10.1111/j.1600-0765.1998.tb02303.x. [DOI] [PubMed] [Google Scholar]
- 42.Albandar JM, Brunelle JA, Kingman A. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J Periodontol. 1999;70:13–29. doi: 10.1902/jop.1999.70.1.13. [DOI] [PubMed] [Google Scholar]
- 43.Bailey MT, et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun. 2010;78:1509–19. doi: 10.1128/IAI.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawley TD, et al. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–9. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cadotte M, Dinnage R, Tilman GD. Phylogenetic diversity promotes ecosystem stability. Ecology. 2012 [Google Scholar]
- 47.Zhang Y, Chen HYH, Reich PB. Forest productivity increases with evenness, species richness and trait variation: a global meta-analysis. Journal of Ecology. 2012;100:742–749. [Google Scholar]
- 48.Petchey O, Gaston K. Effects on ecosystem resilience of biodiversity, extinctions, and the structure of regional species pools. Theoretical Ecology. 2009;2:177–187. [Google Scholar]
- 49.Loreau M, et al. A new look at the relationship between diversity and stability. In: Loreau M, Naeem S, Inchausti P, editors. Biodiversity and Ecosystem Functioning. Synthesis and Perspectives. Oxford University Press; Oxford: 2002. pp. 79–91. [Google Scholar]
- 50.Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- 51.Haak W, et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–9. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 55.Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.