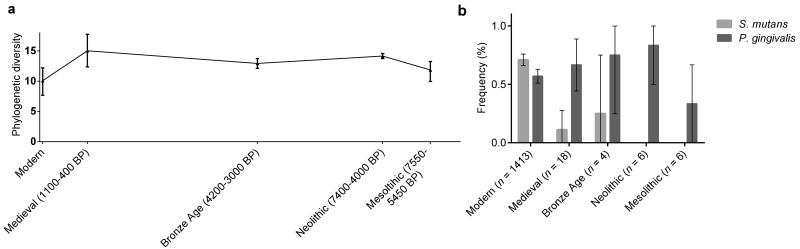
Figure 3. Changes in the diversity and composition of oral microbiota.
(a) For the V3 region sequences, we estimated the phylogenetic diversity50 (Supplementary Note) of the archaeological dental calculus samples (n = 34) and compared them to modern calculus (n = 6) and plaque (n = 13). We estimated phylogenetic diversity from only classified, Gram-positive bacterial sequences to minimize the influence of taphonomic bias (see Supplementary Note). Diversity was calculated at a depth of 34 sequences and bootstrapped to assess the robustness of the pattern. Error bars represent bootstrapped frequencies generated by sampling 255 replicates without replacement. (b) Specific primers were used to amplify sequences unique to the oral pathogens S. mutans (light) and P. gingivalis (dark).