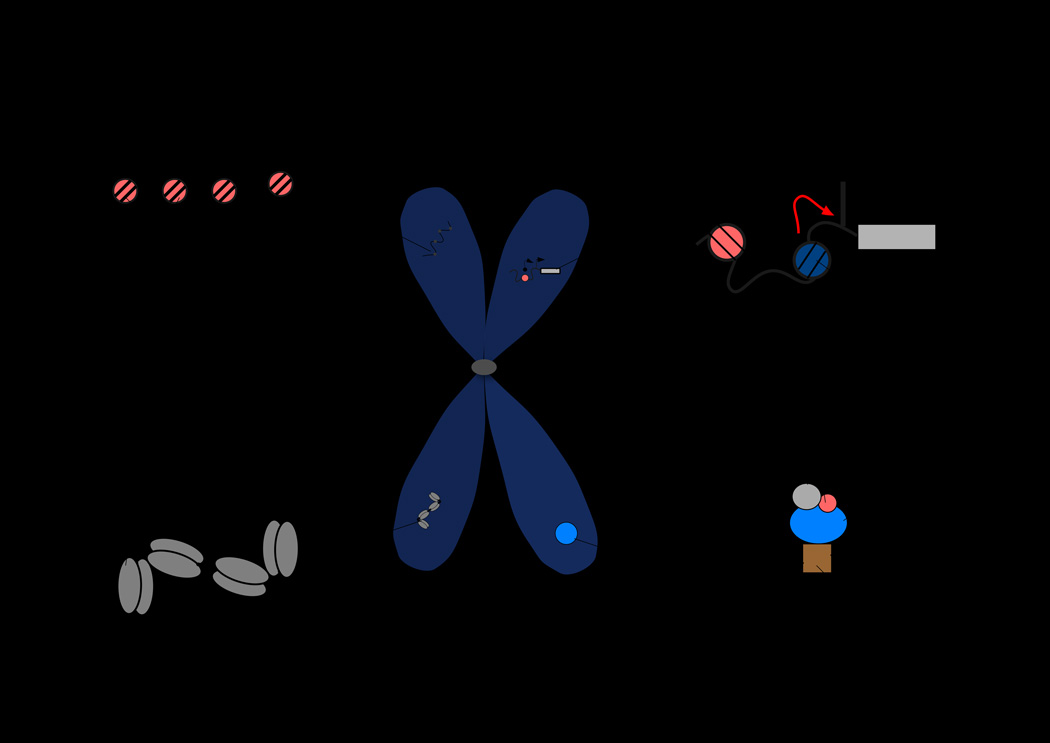
Figure 1. Distinct epigenetic regulatory microenvironments for gene bookmarking that are present in mitotic chromosomes.
Emerging evidence suggest that mitotic chromosomes exhibit distinct epigenetic regulatory microenvironment where various mechanisms of gene bookmarking are operative. These include A) incorporation of histone H3.3 variant into genes that are bookmarked for reactivation post-mitotically, B) `incorporation of histone H2A.Z into nucleosome closer to transcription start site (TSS) and its sliding on to TSS prior to mitosis for post-mitotic expression, C) maintenance of nuclease accessible regions by cohesin where transcription factors are recruited after cell division, and 4) target gene occupancy by sequence transcription factors for post-mitotic gene activation. These microenvironments are not mutually exclusive and may function in concert with other epigenetic mechanisms (e.g., DNA methylation) for maintenance of cellular potential for lineage commitment, growth and proliferation or even disease initiation, progression and perpetuation.