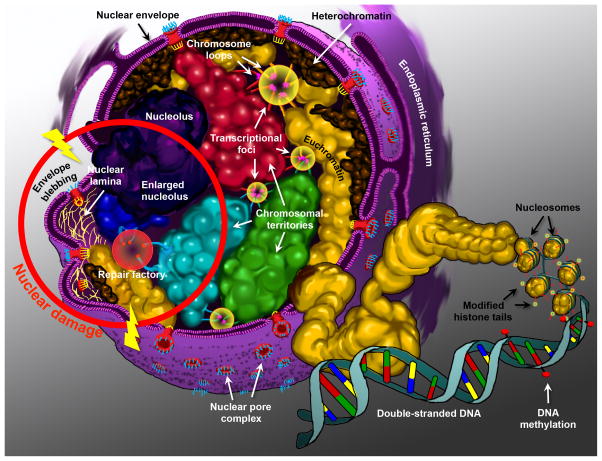
Fig. 2.
Diagram of the nucleus. Highlighting the structural components that contribute to the epigenetic control of genes. Chromosomes are arranged as non-random chromosomal territories (designated by different colors: red, green, cyan, and blue), and are composed of fractal globules of chromosomal folds that establish their ultrastructure. High concentrations of regulatory complexes reside within the interchromosomal space and constitute the nuclear matrix. Highlighted here are the transcriptional foci or factories (yellow circles), where loops of chromosomes can be recruited away from their territories to interact with regulatory elements in trans (red, green, cyan, and gold loops). Chromosomes are also classified into euchromatin (open and more transcriptionally active) (gold domains, plus the red, green, cyan, and blue territories), and heterochromatin (closed and more transcriptionally silent) (brown domains). Heterochromatin is predominantly located at the nuclear periphery. Chromatin is compacted via the winding of DNA around nucleosomes (gold spheroids). Nucleosomes are themselves composed of histone octamers that are epigenetically modified on their histone tails (red and yellow features). DNA methylation (red lollipops) at CpG islands also contributes to the epigenetic control of genes. The nucleolus, the most cytologically prominent feature of the nucleus is illustrated in purple. Perturbations in the nuclear structure that can be attributed to mutations, gene dysregulation, or environmental damage are illustrated within the red ring. Double-strand break repair occurs within repair factories (red circle), which are conducive to translocation events between neighboring chromosomal regions (blue and cyan chromosomes). In laminopathies, the disruption of the nuclear lamina (yellow fibrils) cause nuclear envelope blebbing, and the loss of peripheral localization of certain chromosomal regions. The nuclei of cancer cells are characterized by abnormal, enlarged, and/or fragmented nucleolar morphology.