SUMMARY
Composed of up to 1000 phospho-anhydride bond-linked phosphate monomers, inorganic polyphosphate (polyP) is one of the most ancient, conserved, and enigmatic molecules in biology. Here we demonstrate that polyP functions as a hitherto unrecognized chaperone. We show that polyP stabilizes proteins in vivo, diminishes the need for other chaperone systems to survive proteotoxic stress conditions, and protects a wide variety of proteins against stress-induced unfolding and aggregation. In vitro studies reveal that polyP has protein-like chaperone qualities, binds to unfolding proteins with high affinity in an ATP-independent manner, and supports their productive refolding once non-stress conditions are restored. Our results uncover a universally important function for polyP and suggest that these long chains of inorganic phosphate may have served as one of nature’s first chaperones, a role that continues to the present day.
INTRODUCTION
It is generally agreed that protein synthesis evolved from a world in which RNA served both genetic and catalytic roles in biology, although the driving forces and requirements for the transition to the protein world still remain unclear (Noller, 2004). A major question that has puzzled researchers for a long time is how proteins, which are born as linear chains of amino acids, achieve the intricate three-dimensional structures necessary for proper function. Anfinsen’s classic experiments, which showed that the specific structure of a protein is solely determined by its amino acid sequence, seemed to provide the long-sought answer as to how proteins could have evolved to play such central roles in biology (Anfinsen, 1973). However, it has become increasingly clear that within the crowded environment of the cell, many proteins require a cohort of molecular chaperones, proteases, and regulatory signaling pathways, collectively called the proteostasis network, to fold, function, and withstand stress conditions (Powers and Balch, 2013). This realization raised new questions, particularly regarding the potential coevolution of proteins and the proteostasis mechanisms necessary to keep them stable and soluble. We have now identified a primordial member of the proteostasis network, the prebiotic molecule inorganic polyphosphate (polyP). Synthesized in vivo from ATP and consisting entirely of high-energy phospho-anhydride-bonded inorganic phosphate (Achbergerova and Nahalka, 2011; Rao et al., 2009), these universally conserved molecules exhibit all of the characteristics of an efficient protein chaperone, making polyP one of the most ancient chaperones known.
Chaperone discovery is difficult. Chaperone-deficient cells exhibit many different, seemingly unrelated, and often overlapping phenotypes. These pleiotropic phenotypes are the result of the involvement of molecular chaperones in the folding, assembly and disassembly, transport, and degradation of a large number of different proteins. Therefore the loss of a chaperone can often lead to unpredictable functional effects in the cell (Kim et al., 2013; Powers and Balch, 2013). Compounding this problem, the in vitro assays for chaperones are neither specific nor sensitive enough to enable their purification from crude lysates by activity. It is not surprising, therefore, that new chaperones continue to be discovered even in very well-characterized organisms like Escherichia coli (Quan et al., 2011).
Cells deficient in polyP show a multitude of different phenotypic traits, similar to the pleiotropic phenotypes exhibited by chaperone-deficient cells. Bacteria or unicellular eukaryotes lacking polyP are sensitive to a number of different stress conditions, including heat shock and heavy metal exposure, and are defective in virulence, biofilm formation, and motility (Docampo et al., 2010; Rao et al., 2009). In higher eukaryotes, polyP is known to play a central role in blood clotting, and is involved in apoptosis, mTOR activation, and neuronal signaling (Holmstrom et al., 2013; Kulakovskaya et al., 2012; Moreno and Docampo, 2013; Smith et al., 2010). The underlying physiological role of polyP has been attributed to diverse functions of the molecule: phosphate and energy storage (polyP is isoenergetic to ATP), metal chelation, pH buffering, and regulatory interactions (Kornberg et al., 1999; Kulakovskaya et al., 2012; Rao et al., 2009). However, there is no satisfactory explanation for a general mechanism by which polyP affects these seemingly unrelated processes in the cell.
Here, we show that bacteria, in response to protein-unfolding oxidative stress (i.e., hypochlorous acid, HOCl), re-direct cellular ATP to polyP, resulting in a more than 10,000-fold increase in stress resistance. We demonstrate that polyP functions as a global, highly effective, and wholly inorganic chaperone which stabilizes unfolding proteins, prevents protein aggregation both in vitro and in vivo, and maintains proteins in a refolding-competent form. These results help to explain the long known but largely unexplained role of polyP in protecting organisms against stress conditions, and suggests that polyP may have served as one of nature’s first chaperones.
RESULTS
Phosphate Starvation is an Immediate Response to Oxidative Protein Unfolding Stress
Reexamination of a recent microarray analysis of E. coli gene expression changes in response to the proteotoxic oxidant HOCl, a potent physiological antimicrobial, showed that the expression of at least 12 phosphate starvation-induced genes is highly upregulated (Gray et al., 2013b). This result was consistent with our earlier studies, which revealed that HOCl-treated cells substantially increase their toxic methylglyoxal production (Gray et al., 2013b), a reaction that is driven by low phosphate and high triose phosphate concentrations, and used to restore inorganic phosphate pools (Figure S1A, available online) (Booth et al., 2003). As expected, mutant bacteria carrying deletions in the enzymes DkgA or YqhD, which detoxify the accumulating electrophile methylglyoxal (Figure S1A), were found to be very sensitive to HOCl treatment (Figure 1A). Surprisingly, however, E. coli and Vibrio cholera mutants lacking the enzyme that makes the toxic electrophile (i.e., methylglyoxal synthase, MgsA) were also highly HOCl sensitive (Figure 1A, S1B). These results strongly imply that replenishment of cellular phosphate pools plays a crucial role in oxidative stress defense in bacteria. This was very intriguing since phosphate is known to be entirely non-reactive with reactive chlorine species, and its cellular levels should therefore not be affected by these oxidants (Deborde and von Gunten, 2008).
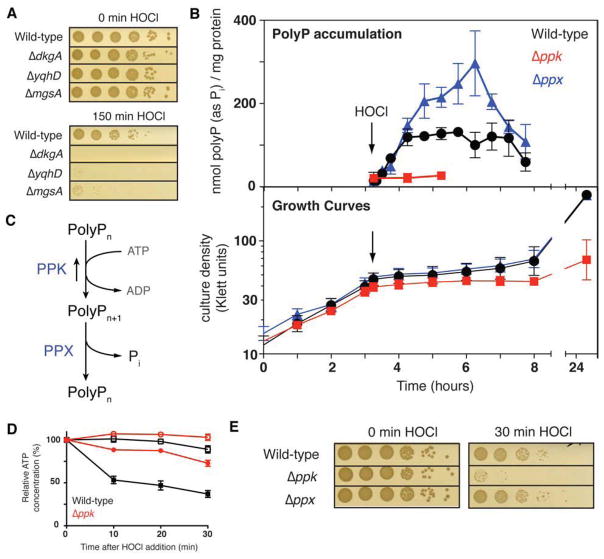
Figure 1. ATP-Derived PolyP Protects Against HOCl.
(A) Exponentially growing E. coli wild-type and mutant strains were incubated with 2 mM HOCl for the indicated time, serially diluted, and spot-titered. See also Figures S1A and S1B.
(B) Growth (lower panel) and intracellular polyP concentration (upper panel) of E. coli wild-type (black circles), Δppk (red squares), and Δppx (blue triangles) (mean ± SD). Arrow indicates addition of 1 mM HOCl. See also Figures S1C, S1D, and S1E.
(C) PolyP synthesis and degradation pathway in E. coli.
(D) ATP content of log phase E. coli wild-type (black squares) and Δppk (red circles) with (closed symbols) or without (open symbols) treatment with 1 mM HOCl, as a percentage of the initial value for each sample (mean ± SD). See also Figure S1F.
(E) Exponentially growing E. coli wild-type and mutant strains were incubated with 2.5 mM HOCl, then diluted and spot-titered. See also Figures S1G and S1H.
Severe Oxidative Stress Leads to PolyP Accumulation
Because phosphate constitutes the building block of polyP, we considered whether polyP accumulation was triggered by HOCl-stress, thereby resulting in phosphate starvation. As shown in Figure S1C, microscopic examination of HOCl-treated E. coli cells stained with 4′-6-diamidino-2-phenylindole (DAPI) revealed a significant accumulation of yellow fluorescent foci, characteristic of DAPI bound to polyP (Aschar-Sobbi et al., 2008). Similar polyP-containing bodies (i.e., “metachromatic granules”, “volutin granules” etc.) have been observed in many stress-exposed organisms, but the purpose of such bodies still remains largely unclear (Docampo et al., 2010; Rao et al., 2009). Very few yellow foci were detected in untreated cells or, as has previously been observed, in cells treated with the non-proteotoxic oxidant hydrogen peroxide (Figure S1C) (Ault-Riche et al., 1998; Winter et al., 2005). Gel analysis (Figure S1D) and quantitative polyP measurements (Figure 1B, S1E) confirmed these results, and showed that E. coli and V. cholerae cells accumulate substantial and comparable amounts of polyP within minutes after HOCl treatment. These results likely provide the missing link between HOCl-stress and phosphate starvation.
PolyP is synthesized from ATP by polyphosphate kinase (PPK). While this is a reversible process, the majority of polyP is degraded to inorganic phosphate by exopolyphosphatase (PPX) under physiological conditions (Figure 1C). Significant polyP accumulation upon HOCl stress should therefore result in a noticeable decline in cellular ATP levels, and would explain why HOCl-stressed organisms appear phosphate starved. Intriguingly, all organisms investigated so far have been shown to experience a very rapid decline in cellular ATP levels upon severe oxidative stress treatment (Barrette et al., 1987; Hyslop et al., 1988; Winter et al., 2005). This ATP decline has previously been attributed to the observed cessation of glycolysis, triggered by the oxidative inactivation of glyceraldehyde-3-phosphate dehydrogenase (GapDH), and leading to the stress-mediated accumulation of dihydroxyacetophosphate (DHAP), the phosphate donor and substrate of methylglyoxal synthase (Figure S1A). In light of our results, however, we now wondered if accumulation of polyP might contribute to the observed ATP decline. To test this idea, we treated wild-type E. coli and the Δppk mutant strain, which is no longer able to respond to HOCl-treatment with the accumulation of polyP (Figures 1B, S1D), with HOCl and measured intracellular ATP levels. Upon exposure to HOCl, wild-type E. coli showed the previously observed rapid decrease in ATP (Winter et al., 2008), while Δppk mutant cells maintained most of their ATP levels (Figure 1D). Comparison of the absolute ATP levels measured in both strain backgrounds (Figure S1F) revealed that within the first 20 min of HOCl-treatment, ATP levels decreased from 1.7 μM ATP/OD600 to 0.8 μM ATP/OD600 in wild-type cells, whereas Δppk strains, which lost significant viability during this time frame (Figure 1E), only showed a decrease in ATP levels from 2.4 μM ATP/OD600 to 2.1 μM ATP/OD600 (Figure S1F). These results suggest that wild-type bacteria actively re-direct a substantial proportion of their cellular ATP pool to form polyP upon exposure to severe oxidative stress, and raise the intriguing possibility that the loss of ATP, long assumed to be a symptom of oxidative stress, may actually be part of an adaptive oxidative stress response aimed at rapidly accumulating large quantities of polyP. It is worth mentioning that the Δppk mutant cells contain substantially higher ATP concentrations than wild-type cells, even under non-stress conditions (Figure S1F). While the implications of this result remain unclear, it is possible that polyP synthesis and degradation act as an ATP-draining futile cycle in wild-type cells, that the presence of polyP stimulates ATP consumption by other enzymes, or that the absence of polyP slows cellular metabolism and/or ATP consumption.
PolyP Accumulation Plays a Critical Role in HOCl defense
To directly test whether accumulation of polyP contributes to the HOCl-resistance of bacteria, we compared the HOCl resistance of E. coli and V. cholerae wild-type cells with strains that lack polyP (i.e., Δppk strains) or that overaccumulate polyP (i.e., Δppx strains). As shown in Figures 1E, S1G and S1H, Δppk cells are exquisitely sensitive to treatment with either HOCl or N-chlorotaurine, a physiologically relevant in vivo secondary oxidation product of HOCl that is present at high concentration in neutrophils (Nagl et al., 2000). In contrast, strains lacking the polyP-degrading exopolyphosphatase PPX (i.e., Δppx strains) accumulate higher levels of polyP upon HOCl-treatment and are slightly more resistant to reactive chlorine species than wild-type cells (Figures 1E, S1H). These results are in agreement with prior studies from Arthur Kornberg’s laboratory and others, showing that polyP-deficient organisms exhibit increased sensitivity towards a variety of environmental stress conditions, including amino acid starvation, osmotic stress, and heat shock (Crooke et al., 1994; Kornberg et al., 1999; Rao et al., 2009).
PolyP Functions as a General Molecular Chaperone In Vivo
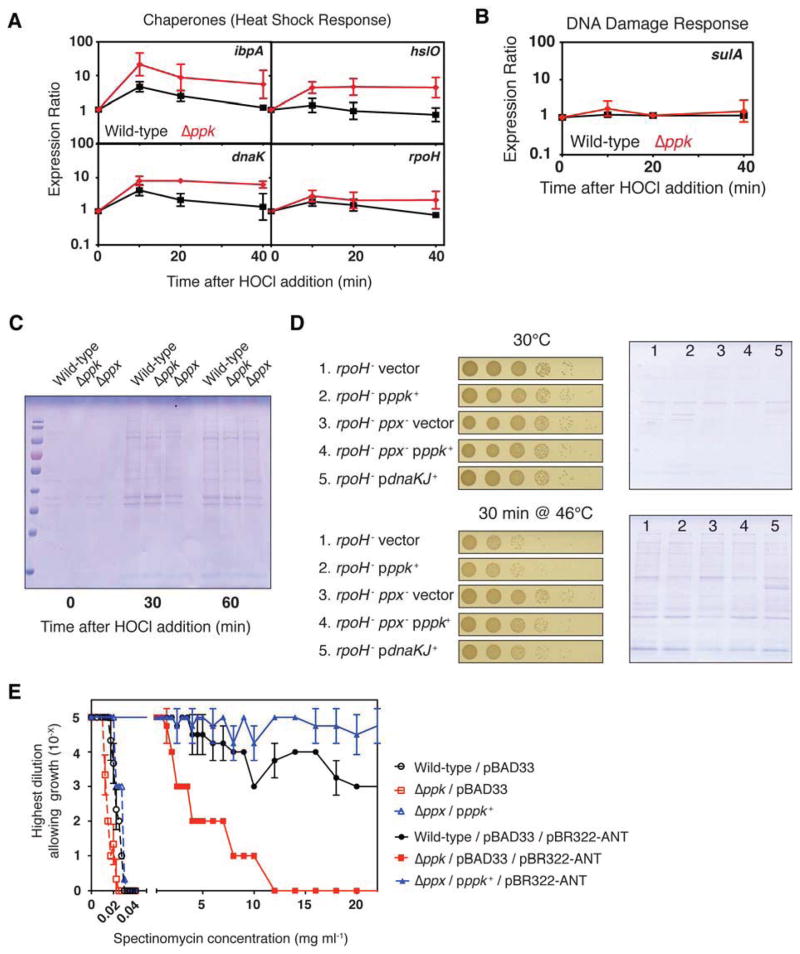
Many stress conditions, including those shown to necessitate polyP accumulation in bacteria for improved survival, cause protein unfolding and aggregation (Kornberg et al., 1999; Winter et al., 2008). As a first test of whether polyP might be involved in maintaining proteostasis, we monitored the expression of heat shock genes in HOCl-treated wild-type and Δppk strains. Stress-induced protein unfolding is the primary trigger of the E. coli heat shock response, making up-regulation of heat shock gene expression a bellwether for the collapse of proteostasis (Guisbert et al., 2008). In the absence of stress, there was no discernable difference in heat shock gene expression in the wild-type and Δppk strains (Figure S2A). However, upon exposure of both strains to HOCl treatment, we found that most of the tested heat shock genes, including the heat shock regulated chaperones Hsp33 and DnaK, were more highly upregulated in Δppk as compared to wild-type (Figure 2A). Expression of rpoH, which encodes the heat shock sigma factor, was also slightly increased. In contrast, marker genes for DNA damage (e.g., sulA), often cited as a major threat during oxidative stress conditions (Imlay, 2013), did not show any additional up-regulation in E. coli Δppk strains (Figure 2B). These results suggest that the presence of polyP reduces the need for other chaperones to combat oxidative protein unfolding, implying that polyP might function as a physiologically relevant chaperone.
Figure 2. PolyP is a Protein-Protective Chaperone In Vivo.
(A) and (B) Wild-type (black) and Δppk (red) E. coli were grown to log phase, then treated with 0.4 mM HOCl. Expression of (A) the heat shock genes ibpA, hslO, dnaK, rpoH, or
(B) the DNA damage indicator gene sulA was measured by qRT-PCR (mean ± SD). See also Figure S2A.
(C) Insoluble protein fractions from exponentially-growing E. coli wild-type and mutant strains before and after addition of 1 mM HOCl.
(D) Survival of (left panels) and insoluble protein fractions from (right panels) exponentially-growing E. coli rpoH− strains before and after a shift from 30° to 46°C.
(E) E. coli strains containing no polyP (red squares), wild-type (black circles), or higher than normal levels of polyP (blue triangles) were grown to log phase, serially diluted, spot-titered on agar containing different concentrations of spectinomycin and scored for growth (mean ± SD). See also Figures S2B and S2C.
To directly test this idea, we treated E. coli wild-type, Δppk, and Δppx strains with HOCl and analyzed the extent of protein aggregation in these cells. As previously observed (Winter et al., 2008), HOCl treatment leads to substantial accumulation of insoluble proteins in wild-type E. coli within 30 min of incubation (Figure 2C). However, cells lacking ppk accumulated slightly more insoluble protein after HOCl treatment than the wild-type whereas cells lacking ppx, and therefore the ability to degrade accumulating polyP, showed substantially less protein aggregation (Figure 2C). These results suggest that polyP plays an important role in protecting cells against toxic protein aggregation, and that levels of polyP accumulation might be inversely correlated with intracellular protein damage.
To test whether this protein-protective effect was specific to HOCl stress conditions or was of general significance, we took advantage of the temperature sensitive rpoH-deficient E. coli strain BB7224. This strain is unable to induce the heat shock response, is largely deficient in protein chaperones, and is extremely sensitive to the protein unfolding effects of heat treatment (Guisbert et al., 2008; Tomoyasu et al., 2001). Overexpression of select chaperones, such as the DnaK/DnaJ system, has been shown to mitigate heat-induced protein aggregation and rescues the temperature sensitive phenotype of this strain (Tomoyasu et al., 2001). We therefore reasoned that, if polyP exerts general chaperone-like protein protection, overproduction of polyP should protect this strain against heat shock in a way comparable to protein chaperones. Since heat stress does not induce significant accumulation of polyP in vivo (Ault-Riche et al., 1998), presumably because the RpoH-dependent proteostasis machinery is fully functional under those stress conditions, we constructed E. coli strains that had been previously shown to contain different levels of polyP (Crooke et al., 1994). We generated BB7224 derivatives with and without functional PPX, containing plasmids expressing ppk (pppk+) from arabinose-inducible promoters. All BB7224 derivatives engineered to overproduce polyP showed substantial improvements in heat shock survival (Figure 2D, left panels) and decreased levels of insoluble protein aggregates (Figure 2D, right panels). The results were comparable to those seen in cells overproducing DnaK/DnaJ (Figure 2D) (Tomoyasu et al., 2001), strongly indicating that polyP plays a direct role in maintaining protein homeostasis in vivo.
To test the ability of polyP to stabilize proteins even under non-stress conditions, we exploited the recent observation that the level of a strain’s antibiotic resistance can be used as read-out for the in vivo stability of the antibiotic resistance protein (Foit and Bardwell, 2013). We therefore co-expressed various antibiotic resistance proteins in wild-type, Δppk, or Δppx pppk+ strains and tested for antibiotic resistance. We observed by far the largest effect of polyP on the spectinomycin resistance-conferring enzyme aminoglycoside 3′-adenyltransferase (ANT), which showed significantly higher antibiotic resistance in PPK-overexpressing strains than in the wild-type, and much lower antibiotic resistance in strains lacking polyP (Figure 2E). Only a very small decrease in spectinomycin resistance was observed for the polyP-deficient strain in the absence of the ANT gene, indicating that the observed effect is largely due to the effect of polyP on ANT. Some apparent stabilization was also seen for chloramphenicol acetyltransferase, but not for the ampicillin resistance protein β-lactamase, which is expressed in the periplasm of E. coli (Figures S2B and S2C). These results demonstrate that polyP acts to stabilize cytoplasmic proteins in vivo, and effectively protects bacteria against stress conditions that cause protein unfolding and aggregation.
PolyP Functions as a General Molecular Chaperone In Vitro
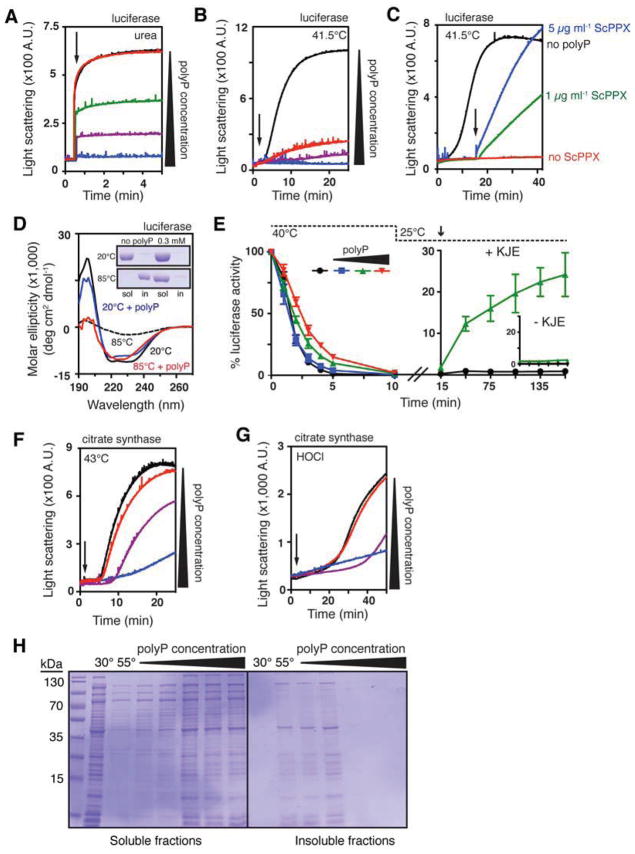
Although our studies demonstrated that polyP works well in a strain background that lacks most chaperones and proteases, the possibility still remained that the observed in vivo effects of polyP are indirect and mediated by a potential influence of polyP on components of the proteostasis machinery. We therefore decided to directly test whether polyP, like a true protein chaperone, recognizes and binds in vitro unfolding proteins and prevents their irreversible aggregation using standard in vitro chaperone assays. We first analyzed the influence of polyP on the aggregation of a variety of previously established chaperone substrate proteins. One such substrate, luciferase, aggregates rapidly when diluted from the urea-denatured form into buffer (Figure 3A, black trace). Astonishingly, micromolar concentrations of polyP (expressed in terms of total phosphate concentration due to the heterogeneous nature of commercially available polyP (Ault-Riche et al., 1998)) inhibited the aggregation of chemically denatured luciferase, with 500 μM polyP completely abolishing luciferase aggregation (Figure 3A, blue trace). PolyP was even more effective in protecting luciferase against thermal aggregation, which typically occurs when luciferase is incubated at temperatures above 40°C (Figure 3B, black trace). The presence of 1 μM polyP was sufficient to significantly reduce thermal aggregation and 100 μM polyP completely prevented aggregate formation (Figure 3B, compare red and blue lines). To determine whether the observed chaperone activity is indeed polyP-specific, and not mediated by additional components in our polyP preparations, we added the highly active, polyP-degrading exopolyphosphatase from Saccharomyces cerevisiae (ScPPX) (Wurst and Kornberg, 1994) to pre-formed polyP-luciferase complexes at elevated temperatures. We reasoned that any luciferase that was subsequently released because of polyP degradation should rapidly aggregate under these conditions. Indeed, addition of ScPPX, which has no discernable chaperone activity or aggregation tendency itself (Figure S3A), resulted in immediate, dose-dependent aggregation of luciferase (Figure 3C, blue and green lines). These results showed that polyP is indeed the chaperone-active component in this assay.
Figure 3. PolyP is a Protein-Protective Chaperone In Vitro.
(A) Aggregation of urea-denatured luciferase upon its dilution (arrow) into buffer containing 0 (black), 0.5 (red), 5 (green), 50 (purple), or 500 μM (blue) polyP. PolyP concentrations are expressed in terms of concentration of inorganic phosphate equivalents (Pi).
(B) Thermal aggregation of luciferase upon its dilution into pre-warmed buffer (arrow) containing 0 (black), 1 (red), 10 (purple), or 100 μM (blue) polyP.
(C) Thermal aggregation of luciferase upon its dilution into pre-warmed buffer containing 0 (black) or 0.5 mM polyP (blue, green, red) and 50 μM MgCl2. Arrow indicates addition of 5 (blue) or 1 (green) μg ml−1 ScPPX. See Figure S3A for additional controls.
(D) Circular dichroism spectra of luciferase incubated with or without 0.3 mM polyP at 20° or 85°C. Inset shows SDS-PAGE of soluble and insoluble luciferase fractions after 20 min incubation at the indicated temperature and spectrum determination.
(E) Left panel: thermal inactivation of luciferase with 0 (black), 0.1 (blue), 1 (green), or 10 mM polyP (red) at 40°C. Right panel: reactivation of luciferase thermally inactivated in the absence (black) or presence (green) of 1 mM polyP upon shift to permissive temperatures and addition of DnaK, DnaJ, GrpE, and MgATP (KJE). Inset shows reactivation in the absence of KJE. Error bars indicate mean ± SD.
(F) Thermal or
(G) HOCl-induced aggregation of citrate synthase with 0 (black), 1 (red), 10 (purple), or 100 mM (blue) polyP. Arrow indicates time of citrate synthase addition (in F) or HOCl addition (in G).
(H) Crude lysates of E. coli ppk::kan+ were incubated 30 minutes at 30° or 55°C, with 0, 0.2, 1, 2, 10, or 20 mM polyP. Soluble and insoluble fractions were separated and examined by SDS-PAGE. See Figure S3B for results with wild-type lysates.
The remarkable ability of polyP to stabilize proteins became clearly apparent when we compared the thermal stability of luciferase in the absence and presence of polyP. While luciferase is completely insoluble upon incubation at 85°C in the absence of polyP (Figure 3D, inset) and has no discernable secondary structure (Figure 3D, black dotted trace), presence of polyP maintained luciferase in a fully soluble and highly structured form for at least 20 min at these near-boiling temperatures (Figure 3D, red trace). These results suggested that polyP keeps luciferase soluble by stabilizing its secondary structure elements. Consistent with this protein-stabilizing effect, incubation of luciferase with increasing amounts of polyP also increasingly delayed the thermal inactivation of luciferase (Figure 3E, left). Quite unexpected was the finding, however, that like a true protein chaperone, polyP maintains thermally inactivated luciferase in a state that is competent for refolding by the DnaK/DnaJ/GrpE system This result became evident when we diluted luciferase, which had been thermally inactivated either in the absence or presence of polyP, into 25°C buffer containing the DnaK/DnaJ/GrpE ATP-dependent chaperone system. Whereas no significant refolding of luciferase was detected in the sample that lacked polyP during the inactivation (Figure 3E, right inset), significant refolding was achieved when luciferase was heat-treated in the presence of polyP (Figure 3E, right). These effects of polyP are very comparable to the effects observed with general protein chaperones like Hsp33 or the small heat shock proteins, which bind unfolding proteins during heat-inactivation and transfer their clients to the DnaK/DnaJ/GrpE chaperone system upon temperature shift for refolding (Haslbeck et al., 2005; Hoffmann et al., 2004; Mogk et al., 1999).
Importantly, polyP’s protein-protective effects were not restricted to thermally or chemically unfolded luciferase but extended also to heat- or HOCl-induced protein aggregation of citrate synthase, another commonly used chaperone substrate. For both thermally or HOCl-unfolding citrate synthase, increasing amounts of polyP in the incubation reaction increasingly prevented protein aggregation (Figures 3F and 3G). To obtain a general overview of the proteins that are protected by polyP and detect any potential client specificity, we added increasing amounts of polyP to crude extracts of E. coli ppk::kan+ (which expresses neither PPK nor PPX) or wild-type and incubated the cell lysates at heat shock temperatures. This strategy has been extensively used to detect clients of protein chaperones. It is based on the observation that most of the proteins that aggregate upon stress treatment in intact cells also aggregate in stress-treated cell lysates (Tomoyasu et al., 2001). We found that addition of polyP broadly protected a large range of different proteins against thermal aggregation and maintained them in a soluble form (Figures 3H and S3B). Importantly, the polyP concentrations necessary to protect bacterial proteins ex vivo (2 – 20 mM) were very similar both to the concentrations required to protect citrate synthase from aggregation in vitro (Figures 3F and 3G) and to the ~ 50 mM polyP concentrations that have been measured in stressed E. coli cells (Ault-Riche et al., 1998). These results strongly suggest that polyP is a promiscuous and general protein chaperone.
PolyP Chain Length Determines its Chaperone Efficacy
The length of polyP chains varies dramatically in nature, and depends both on the organism and the cell type (Rao et al., 2009). While functional differences have been noted between long- and short-chain polyPs, the reasons for these differences are unclear (Smith et al., 2010). To assess whether chain length affects the ability of polyP to protect against protein aggregation, we analyzed the influence of the same concentration (5 mM, based on Pi units) of different defined-length polyPs on the aggregation of thermally unfolding proteins in bacterial cell lysates. We tested the effects of homogenous preparations of 14-mers, 60-mers, and 130-mers, as well as of commercially available heterogeneous mixtures of short-chains of polyP (which were used for all other experiments except where indicated, average: 45 Pi units) and long-chain polyP polymers (range: 200 – 1,300 Pi units). At this concentration, all of the tested polyP preparations exerted some degree of protein protection (Figure 4A). However, by far the most effective chaperones were the long polyP chains, consisting of either 130-mers or a mixture of long-chain polyP (Figure 4A, right-most two lanes). Both preparations almost completely prevented protein aggregation in E. coli cell lysates heated to 55°C. Similar results were obtained when we tested the effect of different chain length polyPs on the aggregation of urea-denatured citrate synthase or thermally-denatured luciferase in vitro. At the minimal concentration of defined-length polyP that effectively protected against aggregation, calculated either in terms of Pi units (Figures S4A and S4B) or in terms of polyP chains (Figures 4B and 4C), we observed a clear dependence of polyP chaperone efficacy on chain length: the shorter the chain length, the less effective the protection. These results are consistent with the finding that bacteria preferentially accumulate long-chain polyP (up to 800 Pi units) upon stress conditions (Ault-Riche et al., 1998; Kornberg et al., 1999). Remarkably, 5 nM of 300-mer polyP chains was sufficient to nearly completely protect 130 nM luciferase against aggregation (Figure 4C, red trace). Higher concentrations (50 μM of chains) were required to protect 80 nM citrate synthase. This result may indicate that polyP exerts its chaperone activity differently with different substrates.
Figure 4. PolyP Chain Length Influences Chaperone Activity.
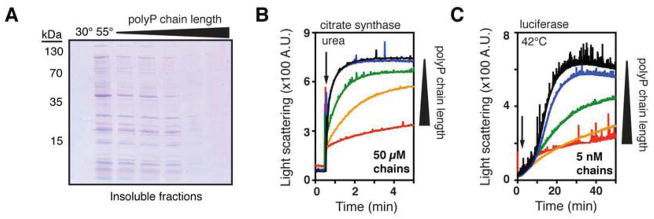
(A) Crude lysates of E. coli ppk::kan+ were incubated 30 minutes at 30° or 55°C, with 0 or 5 mM of different length polyP; from left to right: heterogeneous short-chain (average 45-mer), 14-mer, 60-mer, 130-mer, and long-chain (200–1,300-mer). Insoluble fractions were separated and examined by SDS-PAGE.
(B) and (C) Aggregation of urea-denatured citrate synthase (B) or thermally-denatured luciferase (C) upon their dilution into buffer (arrow) containing no polyP (black) or the indicated concentrations of different length polyP: 14-mer (blue), 60-mer (green), 130-mer (orange), or 300-mer (red). Concentrations were determined according to the length of polyP chains. Corresponding experiments using total Pi concentration are shown in Figures S4A and S4B.
Accumulation of PolyP is Redox-Regulated
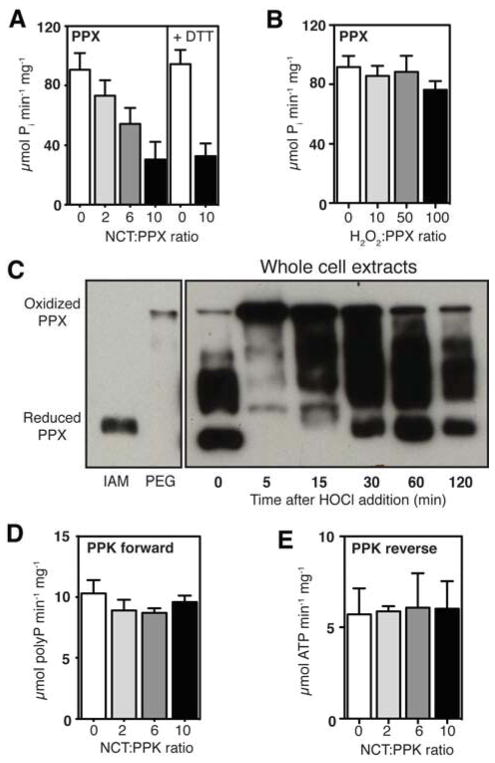
Our in vivo polyP measurements revealed that for the first 60 min of HOCl treatment, Δppx cells accumulate polyP with the same kinetics and to the same extent as HOCl-treated wild-type cells (Figure 1B, top panel, compare blue and black lines). This result was very unexpected since deletion of PPX was predicted to abolish polyP hydrolysis to inorganic phosphate, and therefore should lead to higher levels of polyP in these strains. We therefore considered the possibility that HOCl treatment might transiently inactivate PPX. This would allow for the HOCl-mediated accumulation of polyP, and explain the inorganic phosphate starvation phenotype during HOCl stress. A similar posttranslational regulation has been proposed to trigger polyP accumulation during the E. coli amino acid starvation response. Under these stress conditions, the small signal molecule guanidine 5′,3′-bisdiphosphate (ppGpp) appears to be responsible for the transient inhibition of PPX (Kuroda et al., 1997). However, since HOCl treatment does not induce gene expression changes consistent with ppGpp accumulation (Durfee et al., 2008; Gray et al., 2013b), that mechanism is unlikely to account for HOCl-mediated polyP accumulation. Instead, HOCl is a highly thiol-reactive agent (Gray et al., 2013a), and there is large precedence for organisms using redox-regulated proteins to rapidly mount a stress defense against oxidative protein unfolding stress (Antelmann and Helmann, 2011). We therefore tested the redox sensitivity of PPX in vitro and in vivo. Purified PPX proved to be highly sensitive to inactivation by N-chlorotaurine (which causes many fewer non-specific oxidation artifacts than HOCl (Chapman et al., 2003)) but not by H2O2 (compare Figures 5A and 5B). Mass spectrometric analysis of N-chlorotaurine-treated PPX (Table S1) revealed formation of sulfonic acid, an irreversible thiol modification, on Cys169, which is located in the predicted binding site for polyP (Alvarado et al., 2006), and, to a lesser extent, on the surface-exposed Cys85 (Figure S5A). This result helps to explain not only why oxidation of PPX leads to its inactivation but also why this inactivation is irreversible in vitro (Figure 5A). To monitor PPX oxidation and test for its reversibility in vivo, we conducted differential thiol trapping experiments in E. coli overexpressing PPX (endogenous PPX levels were undetectable with our antibodies) at different time points after HOCl treatment. We alkylated all in vivo reduced cysteines and then labeled all in vivo oxidized cysteines, upon their ex vivo reduction, with the 2 kDa thiol-specific alkylating agent PEG-maleimide. This modification, which indicates the presence of in vivo reversible thiol modifications, leads to significant mass shifts that can be visualized by SDS-PAGE and Western blotting. Within the first 5 min of HOCl treatment, the majority of endogenous PPX shifted to a slower-migrating species, indicating reversible thiol oxidation in PPX (Figure 5C). After about 60 min of HOCl treatment, at least 50% of PPX molecules were again in their reduced state, correlating well with the time at which polyP accumulation begins to level off in wild-type cells while continuing to increase in Δppx cells (Figure 1B). These results provide evidence that PPX is a redox-regulated enzyme, whose HOCl-mediated oxidation and concomitant transient inactivation contributes to the rapid polyP accumulation and Pi depletion in HOCl stressed bacterial cells. Purified PPK, which also contains a cysteine residue in close proximity to its active site (Zhu et al., 2005) (Figure S5B), was not affected by treatment with N-chlorotaurine (Figures 5D and 5E), indicating that the ability of PPK to synthesize polyP and potentially converting it back to ATP is not affected by HOCl treatment.
Figure 5. PPX is a Redox-Regulated Enzyme.
(A) Specific activity of PPX after incubation with different concentrations of N-chlorotaurine (NCT) (mean ± SD). To test for reversibility, oxidatively inactivated PPX (NCT:PPX 10:1) was incubated for 1 hr with 5 mM DTT and assayed again. See also Figure S4A.
(B) Specific activity of PPX after treatment with H2O2 (mean ± SD).
(C) E. coli overexpressing PPX was grown to log phase, then treated with 1 mM HOCl. Reduced cysteine thiols were alkylated with iodoacetamide (IAM). Oxidized cysteine thiols were reduced and alkylated with PEG-maleimide (PEG), adding 2 kDa molecular mass per modified cysteine. PPX was visualized by Western blot. Fully IAM-labeled (“Reduced”) or PEG-maleimide-labeled (“Oxidized”) PPX standards are from the same blot, with longer exposure times.
(D) and (E) Specific activity of PPK’s forward and reverse reactions (see Figure 1C) before and after treatment with NCT (mean ± SD). See also Figure S4B.
DISCUSSION
Here we provide evidence that polyP is an ancient, universally conserved, highly effective, and wholly inorganic protein-protective chaperone, a discovery which may go some way towards explaining the complex pleiotropic phenotypes associated with polyP deficiencies in both prokaryotes and eukaryotes (Docampo et al., 2010; Rao et al., 2009). Our studies in bacteria identified polyP as a key component of a powerful, redox-regulated system for dealing with the proteotoxic effects of fast-acting oxidants such as HOCl. PolyP appears to counteract these proteotoxic effects by stabilizing proteins, preventing irreversible aggregation and maintaining them in a refolding-competent conformation. All these are typical features of protein chaperones, such as the small heat shock proteins or Hsp33. Like protein chaperones, polyP does not appear to have any significant substrate specificity and stabilizes a wide variety of different proteins. While we cannot exclude the possibility that polyP also has additional indirect or regulatory effects on proteostasis in vivo, it is clear from our results that polyP is able to directly stabilize a wide variety of proteins against multiple forms of unfolding stresses.
These results raise the obvious question of how polyP works as a chaperone. Most known chemical chaperones work in a protective osmolyte-like fashion, requiring very high (often molar) concentrations and stabilize proteins via their strong interactions with the solvent (Canchi and Garcia, 2013). In contrast, protein chaperones work in stoichiometric fashion and contain either defined binding sites consisting of a mixture of hydrophobic and charged residues (Kim et al., 2013), or involve intrinsically disordered protein regions, which form upon client binding in a scaffold-like fashion (Kim et al., 2013; Reichmann et al., 2012). PolyP chains are effective at low micromolar concentrations, and their ability to protect proteins against protein aggregation increases with the length of their chain. It is therefore possible that polyP functions as a chemical scaffold, keeping proteins soluble by stabilizing secondary motifs. Alternatively or in combination, ionic interactions between the negatively charged polyP and positive side chains in proteins might contribute to the stabilization effect, as might the high concentration of cations associated with polyP (Kulaev et al., 2004).
Synthesis of polyP does not require transcription or translation. This makes polyP an excellent chaperone during stress conditions, such as HOCl stress, which not only cause protein unfolding but also inhibit new protein translation and inactivate ATP-dependent chaperones, such as the DnaJ/DnaK/GrpE system (Ling and Soll, 2010; Winter et al., 2008). In fact, it is tempting to speculate that synthesis of polyP is part of a larger scheme, in which the high-energy phosphate bonds of ATP are fully preserved during the period while ATP-dependent processes, such as protein translation or ATP-dependent molecular chaperone function, are stalled. This mechanism avoids the costly de novo synthesis of oxidation-prone polypeptide chains and prevents futile cycles between chaperones and unfolding clients under conditions that are non-permissive for folding (Kim et al., 2013). Converting ATP directly into an oxidation-resistant chemical chaperone that binds tightly to and stabilizes unfolding proteins provides immediate compensation for the lack of chaperones. Upon relief of stress, polyP can then be rapidly reconverted to ATP by PPK, restoring cellular energy pools and allowing ATP-dependent chaperones to refold polyP-stabilized proteins (Figure 6). Our findings not only expand the complex redox-regulated network that bacteria use to resist the protein-damaging effects of HOCl (Drazic et al., 2013; Gray et al., 2013b; Winter et al., 2008), but demonstrate a new and fundamentally important function for polyP, one of the most conserved molecules in biology. PolyP’s protein-protective chaperone activity may be key to understanding its fundamental roles and diverse phenotypes in growth, development, virulence, and stress response in both prokaryotes and eukaryotes.
Figure 6. Model of the PolyP Chaperone Cycle.
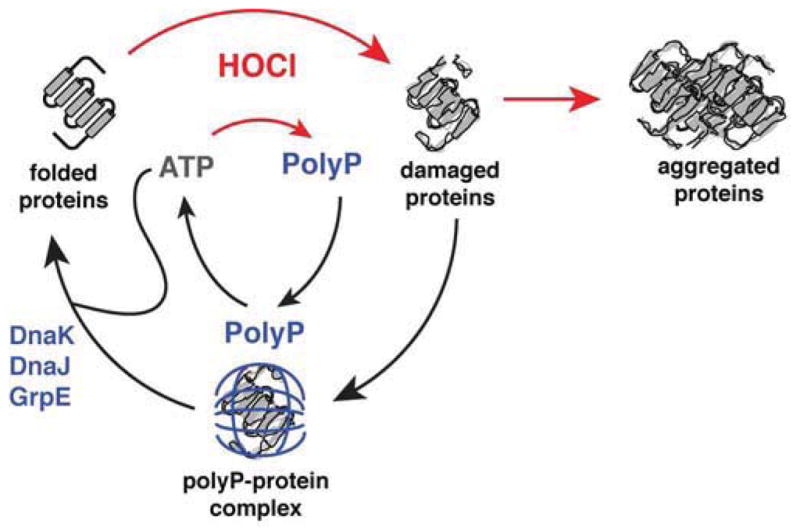
The antimicrobial oxidant HOCl damages proteins, causing formation of cytotoxic protein aggregates. HOCl stimulates rapid conversion of cellular ATP to polyP through the oxidative inactivation of PPX. This conversion conserves high-energy phospho-anhydride bonds while down-regulating cellular processes that require ATP, including ATP-dependent chaperones like DnaK. PolyP functionally replaces these chaperones by forming stable complexes with unfolding proteins, keeping them soluble and refolding-competent. Upon relief of stress conditions, polyP may be either degraded to free phosphate by PPX or reconverted to ATP by PPK. Restoration of cellular ATP pools reactivates ATP-dependent chaperones and allows for the effective refolding of polyP-protected proteins by the DnaK/DnaJ/GrpE complex.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Growth Conditions, and Molecular Methods
All strains and plasmids used in this study are listed in the Table of Strains and Plasmids in the Supplemental Information. Bacterial manipulations, protein purifications, enzyme assays, thiol trapping, and mass spectrometry were carried out according to standard methods; details are available online in Supplemental Experimental Procedures.
Phenotypic and Transcriptional Analyses
HOCl survival assays and qRT-PCR of E. coli were performed as described (Gray et al., 2013b). Survival assays for V. cholerae were the same as for E. coli, except that 550 μM HOCl was used instead of 2 mM HOCl and 2 mM methionine was used to quench HOCl instead of 10 mM sodium thiosulfate (Na2S2O3). For N-chlorotaurine tolerance, E. coli was grown in MOPS medium at 37°C to an OD 600 ~ 0.4, then concentrated by centrifugation and resuspended to an OD600 = 0.1 in the same medium with and without 120 μM N-chlorotaurine. Growth curves were collected using a Bio-Tek Synergy HT plate reader, incubating the cultures at 37°C with constant shaking.
Quantitative High-Throughput PolyP Assay
Intracellular polyP levels were measured according to (Ault-Riche et al., 1998) with slight modifications. A Biomek® FX fluid handling robot (Beckman Coulter) was used to automate polyP extraction, digestion, and measurement; full details are available online in Supplemental Experimental Procedures.
In Vivo Protein Aggregation Assays
Membrane protein-free cellular insoluble protein fractions were prepared by a modification of the method of (Tomoyasu et al., 2001). Briefly, cells equivalent to 4 ml of OD600 = 1 were harvested by centrifugation and resuspended in 40 μl Buffer 1 (10 mM potassium phosphate (pH 6.5), 1 mM EDTA, 20% (w/v) sucrose, 1 mg ml−1 lysozyme, 50 U ml−1 Benzonase nuclease (Merck)), then incubated 30 min on ice and frozen at −80°C. After thawing on ice and addition of 360 μl Buffer 2 (10 mM potassium phosphate (pH 6.5), 1 mM EDTA), cells were transferred to 2 ml microfuge tubes containing ~ 200 μl 0.5 mm glass beads (BioSpec Products) and shaken for 30 min @ 1400 rpm @ 8°C to lyse cells completely. 200 μl aliquots were taken and insoluble fractions were separated by centrifugation (20 min @ 16,100 g @ 4°C), rinsed once with Buffer 2, once with Buffer 3 (Buffer 2 plus 2% Nonidet P-40 (ICN Biomedicals)), and again with Buffer 2, then visualized by reducing SDS-PAGE. For HOCl stress, E. coli strains were grown in MOPS medium at 37°C to OD 600 ~ 1, then diluted to OD600 = 0.35 with fresh medium. HOCl was added to 1 mM and incubation was continued. Samples were taken at the indicated time points, quenching HOCl by immediate addition of Na2S2O3 to 10 mM. For heat stress, BB7224-derived E. coli strains were grown at 30°C in LB medium containing ampicillin and 1% arabinose to OD600 ~ 1, then diluted to OD600 = 0.6 with fresh medium and incubated at 46°C. Samples were taken at the indicated time points, cooled rapidly, and cell survival was assessed at 30°C by serial dilution on LB agar plates containing 1% arabinose.
In Vivo Effect of PolyP on Antibiotic Resistance Protein Stability
E. coli strains MG1655/pBAD33, Δppk/pBAD33, and Δppx/pPPK1 were transformed with pBR322-ANT. Overnight cultures of these strains were diluted 1:100 in fresh LB and grown to log phase at 37°C. OD 600 was normalized to 1 with phosphate buffered saline (PBS). Cells were serially diluted in PBS and 2 μl of dilutions 100 to 10−5 were spotted on LB agar plates containing 1% arabinose and the indicated concentrations of antibiotics.
In Vitro Protein Aggregation Assays
For aggregation of denatured proteins, citrate synthase or luciferase (12 μM) were denatured in urea (6.5 M for luciferase, 7.5 M for citrate synthase) for 2 hours, then diluted to 60 nM (luciferase) or 80 nM (citrate synthase) in 40 mM potassium phosphate (pH 7.5) at 30°C containing polyP as indicated. For thermally-induced protein aggregation, citrate synthase (0.30 μM) was incubated in 40 mM potassium phosphate (pH 7.5) at 43°C or luciferase (0.13 μM) was incubated in 40 mM HEPES (pH 7.5) at 41.5°C – 43°C with the indicated amounts of polyP. For HOCl-induced protein aggregation, citrate synthase (3 μM) was incubated in 40 mM potassium phosphate (pH 7.5) at 30°C with 350 μM HOCl. Light scattering was measured at λex and λem = 360 nm using a Hitachi F4500 fluorescence spectrophotometer with a thermostatted cuvette holder under constant stirring. All experiments were performed at least in triplicate. Each panel shows representative results obtained on a single day with a single batch of protein. 200 μg aliquots of crude cell lysates with polyP added as indicated were incubated 30 min with shaking (650 rpm) at the indicated temperatures. Soluble and insoluble fractions were separated by centrifugation (20 min @ 16,100 g @ 4°C) and visualized by reducing SDS-PAGE.
Thermal Inactivation and Refolding of Luciferase
DnaK, DnaJ, and GrpE were purified as described (Hoffmann et al., 2004). Luciferase (4 μM) was incubated in 10 mM potassium phosphate (pH 7.5) at 40°C, and samples were removed at the indicated time points. After 10 min, samples were transferred to 25°C for 5 min, then diluted 1:40 into 40 mM HEPES (pH 7.5), 50 mM KCl, 0.1 mg ml−1 bovine serum albumin, 2 mM MgATP with or without the addition of 1 μM DnaK, 0.2 μM DnaJ, and 1 μM GrpE. Luciferase activity was determined by luminescence (Lundin, 2000), in reaction mixtures containing 20 nM luciferase, 25 mM Tricine (pH 7.8), 5 mM MgSO4, 0.1 mM EDTA, 1 mM DTT, 35 μM luciferin, and 2 mM MgATP.
Circular Dichroism (CD) Spectroscopy
CD spectra (190–260 nm) were measured for 0.4 mg ml−1 luciferase in 10 mM potassium phosphate (pH 7.5) with or without 0.3 mM polyP at 20°C and 85°C, using a J-810 CD spectrophotometer (Jasco). After measurement, soluble and insoluble protein fractions of each treatment were separated by centrifugation (20 min @ 16,100 g @ 4°C) and visualized by reducing SDS-PAGE.
Supplementary Material
HIGHLIGHTS.
Polyphosphate protects against proteotoxic oxidative stress conditions.
Polyphosphate is a prebiotic and universally conserved molecular chaperone.
Different length polyphosphates differ in their chaperone efficacies.
Polyphosphate accumulation is mediated by the redox-regulated enzyme PPX.
Acknowledgments
We thank Dr. Carol Gross for her extremely helpful advice on preparing this manuscript. We are grateful to Dr. Toshikazu Shiba (Regenetiss, Japan) for providing us with polyphosphates of defined chain length. This work was funded by National Institute of Health grants GM065318 and AI097893 to U.J., F32-GM096613 to M.J.G., and T32-GM007315 to W.Y.W. J.C.A.B. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achbergerova L, Nahalka J. Polyphosphate--an ancient energy source and active metabolic regulator. Microb Cell Fact. 2011;10:63. doi: 10.1186/1475-2859-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado J, Ghosh A, Janovitz T, Jauregui A, Hasson MS, Sanders DA. Origin of exopolyphosphatase processivity: Fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homolog. Structure. 2006;14:1263–1272. doi: 10.1016/j.str.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxidants & redox signaling. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, Pavlov E. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc. 2008;18:859–866. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- Ault-Riche D, Fraley CD, Tzeng CM, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrette WC, Albrich JM, Hurst JK. Hypochlorous Acid-Promoted Loss of Metabolic Energy in Escherichia-Coli. Infect Immun. 1987;55:2518–2525. doi: 10.1128/iai.55.10.2518-2525.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR, Ferguson GP, Miller S, Li C, Gunasekera B, Kinghorn S. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem Soc Trans. 2003;31:1406–1408. doi: 10.1042/bst0311406. [DOI] [PubMed] [Google Scholar]
- Canchi DR, Garcia AE. Cosolvent effects on protein stability. Annu Rev Phys Chem. 2013;64:273–293. doi: 10.1146/annurev-physchem-040412-110156. [DOI] [PubMed] [Google Scholar]
- Chapman AL, Winterbourn CC, Brennan SO, Jordan TW, Kettle AJ. Characterization of non-covalent oligomers of proteins treated with hypochlorous acid. Biochem J. 2003;375:33–40. doi: 10.1042/BJ20030685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E, Akiyama M, Rao NN, Kornberg A. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1994;269:6290–6295. [PubMed] [Google Scholar]
- Deborde M, von Gunten U. Reactions of chlorine with inorganic and organic compounds during water treatment-Kinetics and mechanisms: a critical review. Water Res. 2008;42:13–51. doi: 10.1016/j.watres.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Docampo R, Ulrich P, Moreno SNJ. Evolution of acidocalcisomes and their role in polyphosphate storage and osmoregulation in eukaryotic microbes. Philos T R Soc B. 2010;365:775–784. doi: 10.1098/rstb.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Miura H, Peschek J, Le Y, Bach NC, Kriehuber T, Winter J. Proceedings of the National Academy of Sciences of the United States of America. 2013. Methionine oxidation activates a transcription factor in response to oxidative stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foit L, Bardwell JC. A tripartite fusion system for the selection of protein variants with increased stability in vivo. Methods Mol Biol. 2013;978:1–20. doi: 10.1007/978-1-62703-293-3_1. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Jakob U. Bacterial responses to reactive chlorine species. Annual review of microbiology. 2013a;67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Parker BW, Kim M, Jakob U. NemR is a Bleach-Sensing Transcription Factor. J Biol Chem. 2013b;288:13789–13798. doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nature Structural & Molecular Biology. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. EMBO J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom KM, Marina N, Baev AY, Wood NW, Gourine AV, Abramov AY. Signalling properties of inorganic polyphosphate in the mammalian brain. Nature communications. 2013;4:1362. doi: 10.1038/ncomms2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyslop PA, Hinshaw DB, Halsey WA, Jr, Schraufstatter IU, Sauerheber RD, Spragg RG, Jackson JH, Cochrane CG. Mechanisms of oxidant-mediated cell injury. The glycolytic and mitochondrial pathways of ADP phosphorylation are major intracellular targets inactivated by hydrogen peroxide. J Biol Chem. 1988;263:1665–1675. [PubMed] [Google Scholar]
- Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature reviews Microbiology. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kulaev IS, Vagabov VM, Kulakovskaya TV. The Biochemistry of Inorganic Polyphosphates. Chichester, England: John Wiley & Sons, Ltd; 2004. The chemical structures and properties of condensed inorganic phosphates; pp. 3–13. [Google Scholar]
- Kulakovskaya TV, Vagabov VM, Kulaev IS. Inorganic polyphosphate in industry, agriculture and medicine: Modern state and outlook. Process Biochem. 2012;47:1–10. [Google Scholar]
- Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- Ling J, Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A. Use of firefly luciferase in ATP-related assays of biomass, enzymes, and metabolites. Method Enzymol. 2000;305:346–370. doi: 10.1016/s0076-6879(00)05499-9. [DOI] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SN, Docampo R. Polyphosphate and its diverse functions in host cells and pathogens. PLoS pathogens. 2013;9:e1003230. doi: 10.1371/journal.ppat.1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl M, Hess MW, Pfaller K, Hengster P, Gottardi W. Bactericidal activity of micromolar N-chlorotaurine: evidence for its antimicrobial function in the human defense system. Antimicrob Agents Chemother. 2000;44:2507–2513. doi: 10.1128/aac.44.9.2507-2513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF. The driving force for molecular evolution of translation. Rna. 2004;10:1833–1837. doi: 10.1261/rna.7142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Balch WE. Diversity in the origins of proteostasis networks--a driver for protein function in evolution. Nature reviews Molecular cell biology. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, et al. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol. 2011;18:262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Choi SH, Davis-Harrison R, Huyck J, Boettcher J, Rienstra CM, Morrissey JH. Polyphosphate exerts differential effects on blood clotting, depending on polymer size. Blood. 2010;116:4353–4359. doi: 10.1182/blood-2010-01-266791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Molecular microbiology. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Wurst H, Kornberg A. A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. J Biol Chem. 1994;269:10996–11001. [PubMed] [Google Scholar]
- Zhu Y, Huang W, Lee SS, Xu W. Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep. 2005;6:681–687. doi: 10.1038/sj.embor.7400448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.