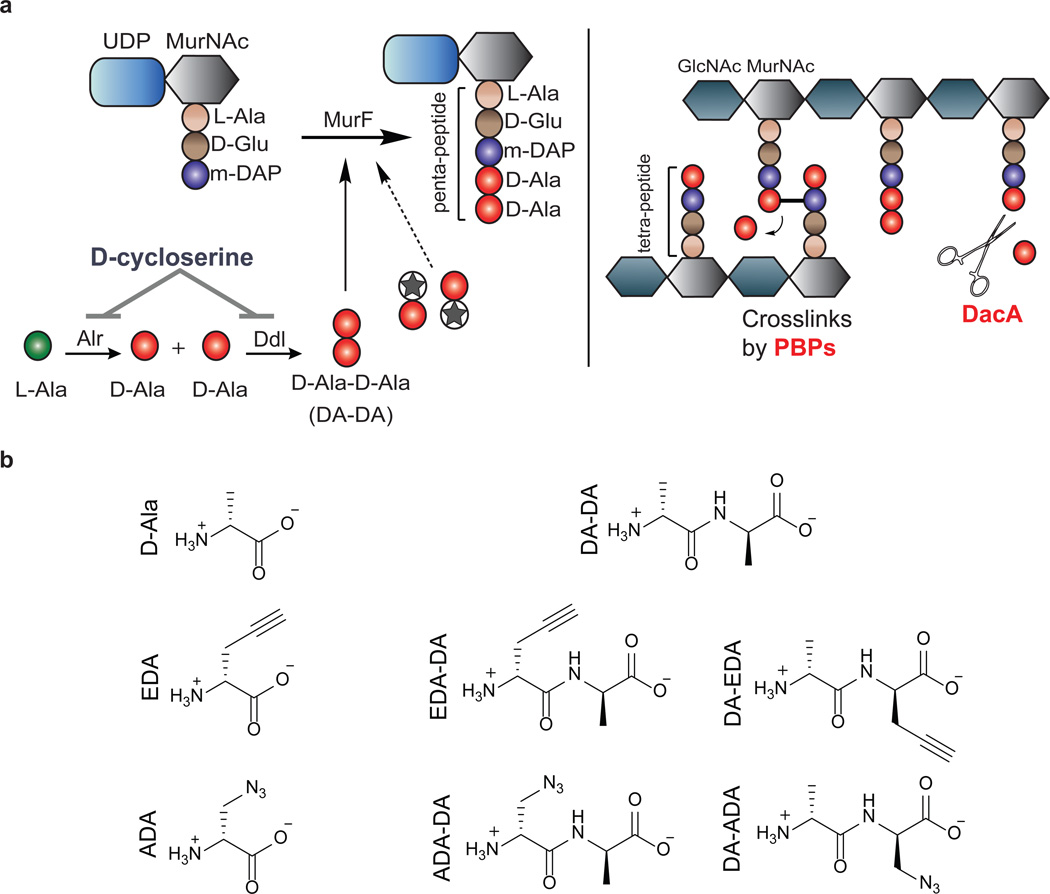
Figure 1. Novel dipeptide PG labeling strategy.
a, Biosynthesis of the terminal PG stem peptide of Gram negative bacteria. Two D-alanines are first ligated together by D-alanine-D-alanine ligase and the dipeptide is subsequently added to the stem tripeptide by MurF, resulting in a pentapeptide. The labeling strategy relies on the inherent tolerance of the PG machinery to accept DA-DA analogs. Subsequent cross-linking between neighboring peptide stems is carried out by a series of transpeptidases (penicillin binding proteins). Upon transpeptidation, a proximal m-DAP from a neighboring peptide stem attacks the subterminal D-alanine of the PG stem. The terminal D-alanine is thus cleaved from the stem peptide, which results in a tetrapeptide. Another pathway for the loss of terminal D-alanine is D,D-carboxypeptidation catalyzed by enzymes such as DacA. b, Chemical structures of D-Ala, DA-DA, and their derivatives carrying bioorthogonal handles used in this study. Abbreviations: MurNAc, N-acetylmuramic acid; GlcNAc, N-acetylglucosamine; L-Ala, L-Alanine; D-Glu, D-Glutamic Acid; m-DAP, meso-diaminopimelic acid; D-Ala, D-Alanine; Alr, Alanine racemase; Ddl, D-alanine-D-alanine ligase; MurF, UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase; PBPs, penicillin-binding proteins; DA-DA, D-alanyl-D-Alanine; EDA, Ethynyl-D-alanine; ADA, Azido-D-alanine; EDA-DA, Ethynyl-D-alanyl-D-alanine; ADA-DA, Azido-D-alanyl-D-alanine.