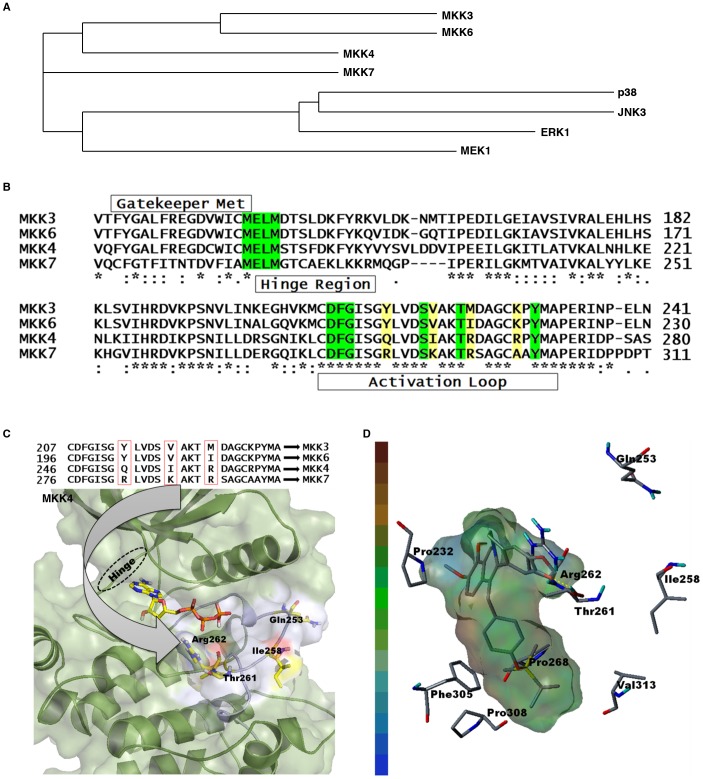
Figure 4. MKK4 and MKK7 homology in mammalian MAPK pathways.
A) Phylogenetic tree for human MKK4, MKK7, MKK3, MKK6, MEK1, p38, JNK, and ERK. B) Amino acid sequence alignment starting from the hinge region to the activation loop are shown for MKK3 (UniProtKB accession code: P46734), MKK6 (UniProtKB accession code: P52564), MKK4 (UniProtKB accession code: P45985), and MKK7 (UniProtKB accession code: O14733). Green denotes a highly conserved region among these MKKs. Residues highlighted in yellow designate sequence variations in the activation loop. C, D) Three-dimensional structure of MKK4 suggests that HWY336 interacts with the activation loop through hydrogen bonding. C) (top) Amino acid sequence variations within the activation loop of MKKs. (bottom) Proposed docked pose of ATP in MKK4. The arrow designates different amino acids that may determine MKK selectivity. The MKK4 structure is shown in the background with the activation loop (white) containing the varying amino acids (Arg262) at the respective positions (generated with the Pymol program; www.pymol.org). D) Hydrophobic interactions between HWY336 and the MKK4 active site are shown. Hydrophobic residues within the active site are designated by the cap-stick model and HWY336 is displayed using transparent hydrophobic surfaces. The hydrophobicity index is displayed on the left, where brown and blue denote highly hydrophobic and hydrophilic areas, respectively. Pro268, Phe305, Pro308, and Val313 interact with HWY336 side chains. HWY336 interacts with the activation loop of MKK4 through hydrogen bonding via the hydroxyl group of Thr261.