Abstract
Porous silica particles are potential transfection agents for nucleic-acid based therapies due to their large specific surface areas and pore volumes and the ease with which they can be chemically modified to maximize the loading of cargo and to effect targeting in vivo. Here, we present a systematic study of the effects of pore size and pore modification on the adsorption and release of short, interfering RNA (siRNA) from a mesoporous silica particle developed in our laboratory. Using adsorption isotherms and release experiments, we found that the short polyamine diethylenetriamine was the best chemical modification for achieving both the adsorption and release of large amounts of siRNA. The degree of functionalization with diethylenetriamine caused drastic changes to the loading capacity and binding strength of siRNA to silica with relatively large pores (8 nm and larger), but degree of functionalization had a weaker effect in narrow pores (4 nm). Multilayer adsorption could occur in materials with large pores (15 nm). Release experiments showed that intermediate pore sizes and intermediate degrees of functionalization resulted in the best compromise between maximizing loading (from strong adsorption) and maximizing release. Capillary electrophoresis and quantitative, real-time PCR demonstrated that the siRNA was released intact and that these particles functioned as a transfection agent to mammalian cells in vitro.
Keywords: mesoporous silica, siRNA, adsorption, transfection agent, drug delivery, Langmuir adsorption, Freundlich adsorption
Introduction
RNA interference (RNAi) is a relatively new concept, confirmed in higher animals only a decade ago,1 and it already holds tremendous promise for gene therapy of a wide-range of diseases. Briefly, RNAi effects the knockdown of specific genes using sequences of short hairpin RNA (shRNA) or short interfering RNA (siRNA) and cells’ endogenous RNAi machinery, the RNA-induced silencing complex (RISC). Because genetic malfunctions underpin so many cancers, RNAi has been most vigorously pursued for treating cancer;2 however, many hurdles must be overcome before siRNA can be used as a therapeutic agent. Unprotected RNA is rapidly degraded in the body by endogenous ribonucleases (RNases) and the negatively charged RNA strands cannot pass through cell membranes.3 Moreover, systemic administration of siRNA can cause immune responses and off-target gene effects.4–6 The simplest way to overcome the poor pharmacokinetics and immunogenicity of siRNA is via chemical modifications of the RNA backbone;7–9 yet, these methods cannot be used to create targeted siRNA-delivery vehicles. An alternative method called electroporation involves focusing an electric field on the tissue of interest to permeabilize cell membranes to naked or modified siRNA, but this approach would be difficult to employ with tumors deep within the body.10,11
Due to these problems with naked RNA, most therapeutic approaches with RNAi use transfection agents to transport the siRNA or shRNA into tissues and cells of interest. Many transfection agents aggregate with or fully encapsulate the RNA, thereby disguising it from the immune system and RNases. One of the most commonly used RNA interference vectors employs the envelopes of modified viruses to deliver DNA cassettes to cells that then synthesize the required siRNAs in situ.12 A variety of virus types can be used and in vivo studies have shown promise,13,14 but the immunogenicity and cytopathic effects of virus-based materials cause concern for widespread use. Chemical transfection agents include synthetic polymers, liposomes, and nanoparticles. Most polymeric transfection agents are cationic polyamines and rely on electrostatic interactions with the anionic siRNA to form stable complexes,15,16 but the toxicity of free polyamines15 limits their use in therapeutic settings. Synthetic nanoparticles, including self-assembled polyplexes,17 liposomes,18–21 porous silica,22–29 quantum dots,30 and metal oxides,31 have been reported for gene therapy in vitro and in vivo. Particle-based transfection agents are attractive because they can be chemically modified with moieties such as biocompatible polymers for improved biodistribution and pharmacokinetics and with peptides or antibodies to effect tissue-specific delivery of siRNA within the body. Porous particles are particularly promising due to their large surface areas and internal pore volumes for loading nucleic acid-based therapies, but few reports have used such materials as transfection agents for siRNA.
Herein, we present studies on the uptake and release of siRNA from porous silica microparticles called acid-prepared mesoporous spheres (APMS)32–35. Using differential functionalization of the particle exteriors and pore surfaces, we modified the particle exteriors with a biocompatible polymer to enable cellular uptake while testing a variety of pore surface modifications and pore diameters to optimize the loading and release of siRNA, as determined through adsorption isotherms and release kinetics. We found that the type and degree of functionalization lead to markedly different adsorptive behavior. Besides representing, to the best of our knowledge, the first systematic study of the effects of pore size and pore modification on the adsorption and release of siRNA from porous silica, our system differs in several ways from previous transfection agents based on porous silica.23–29 Our particles are larger, thus avoiding possible toxicity issues surrounding nanoparticles, such as acute toxicity caused by disruption of normal cell functions36–39 and long-term toxicity caused by bioaccumulation.40–42 Also, they are not membrane-bound upon internalization, avoiding the need for endosomal escape.32 Finally, the siRNA is adsorbed inside the particles rather than on the surface,24,43 protecting the cargo without affecting the cellular uptake of the particles.
Experimental Section
Materials and Methods
Reagents
Unless otherwise noted, chemical and cell culture reagents were purchased from commercial sources and used as received. Triethylamine, toluene, and methylene chloride were distilled prior to use, and acetonitrile (MeCN) was stored over molecular sieves. RNase-free water and/or UV-sterilized growth medium were used for any experiments involving siRNA.
The siRNA used in adsorption and release studies was the AllStars negative control siRNA (Qiagen) with an AlexaFluor®-647 label on the 3′ end of the sense strand, whose sequence was:
Sense GGGUAUCGACGAUUACAAAUU
Antisense UUUGUAAUCGUCGAUACCCUG
Nitrogen Physisorption
N2 adsorption and desorption isotherms were obtained on a Micromeritics TriStar instrument. Surface areas were measured using the BET method and pore size distributions were calculated from a modified KJS method using the adsorption branch.
Confocal Laser Scanning Microscopy (CLSM)
Particles were mounted onto slides using Poly-Mount® (Polysciences, Inc.) and analyzed by CLSM (Carl Zeiss LSM 510 META laser scanning microscope). The AlexaFluor®-647 dye label on the siRNA was excited with a HeNe laser at a wavelength of 633 nm and the particles were imaged with a 63 X oil-immersion objective and acquired using a long-pass 650 filter.
Bioanalyzer
siRNA from a stock solution and that recovered after release from APMS were subjected to capillary electrophoresis using the Agilent 2100 Bioanalyzer. This platform uses a microfluidic “lab-on-chip” design, incorporating capillary electrophoresis and fluorescence, combined with the 2100 Expert software to evaluate both RNA concentration and integrity.
Cell Culture
Small airway epithelial cells (SAEC, Tom K. Hei, Columbia University)44 were grown and maintained in DMEM/F12 50:50 media containing 10% Fetal Bovine Serum (FBS) (CellGro® Mediatech inc, Manassas, VA), with penicillin (50 units/ml), streptomycin (100 μg/ml) (Invitrogen, Carlsbad, CA), hydrocortisone (100 μg/ml), insulin (2.5 μg/ml), transferrin (2.5 μg/ml) and selenium (2.5 μg/ml) (Sigma, St. Louis, MO).
Treatment with Lipofectamine and APMS/siRNA
SAEC cells were plated in 12-well plates and grown for two days until they were roughly 40 % confluent. On the treatment day, the medium was aspirated and replaced with Opti-MEM® (Life Technologies) reduced-serum medium. Cells were treated with either A) APMS that had been previously loaded with siRNA, centrifuged, and rinsed once with RNase-free water or B) siRNA/Lipofectamine™ 2000 (Life Technologies) complexes prepared according to the manufacturers instructions at a ratio of 10 pmol siRNA per μL of transfection agent as previously reported by our collaborators.45 Cells were further incubated for another 24 hr before harvesting for qRT-PCR analysis.
Quantitative, Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was prepared using an RNeasy® Plus Mini Kit according to the manufacturers’ protocol (Qiagen, Valencia, CA), as published previously.45 Then, total RNA (1.0 μg) was reverse-transcribed with random primers using the AMV Reverse Transcriptase kit (Pro-mega, Madison, WI) according to the recommendations of the manufacturer. Taqman® qRT-PCR was performed using Assays-On-Demand™ primer and probe sets (Applied Biosystems, Foster City, CA) by the Vermont Cancer Center Advanced Genome Technologies Core facility.
Synthesis of Modified APMS-(s)TEG Particles
APMS functionalized on its external surface with TEG and with diethylenetriamine in its pores [APMS-DETA-(s)TEG]46
APMS particles were first functionalized exclusively on their external surfaces with tetraethylene glycol, APMS-(s)TEG, as previously described.32,47,48 These particles (150 mg) were then suspended in hexanes (~10 mL) with rapid stirring. To these stirred suspensions were added various volumes of a stock solution of 1-(3-trimethoxysilylpropyl)-diethylenetriamine (TMSP-DETA) in dichloromethane (11.3 mg mL−1) to make a total volume of 10.0 mL, and stirring was continued for ten minutes. The particles were then filtered quickly without rinsing, air dried for 30 minutes, and finally cured at 80 °C overnight.
APMS functionalized on its external surface with TEG and with Mg2+ in its pores [APMS-Mg-(s)TEG]43
The pores were doped with Mg2+ ions by simply soaking APMS-(s)TEG, prepared as previously described,43 in a solution of MgCl2 (100 mM) an hour before the addition of the siRNA. The doped particles were not otherwise isolated.
APMS functionalized on its external surface with TEG and with imidazole in its pores [APMS-Im-(s)TEG]49,50
Briefly, APMS-(s)TEG (2.0 g) was first refluxed in dry toluene (15 mL) with 3-chlorotrimethoxysilane (0.5 g) overnight, then filtered and rinsed with excess toluene. The resulting particles, now with primary chloro groups immobilized exclusively in the pores, were subsequently refluxed in m-xylenes (15 mL) with freshly recrystallized imidazole (0.28 g, 3.5 mmol) overnight. The final particles were rinsed thoroughly with toluene, acetone, and lastly methanol before drying in a 110 °C vacuum oven.
Adsorption and Release of siRNA
siRNA Adsorption
Sub-milligram quantities of APMS particles were first measured into vials by the following method. Particles (~20 mg) were weighed into a 15 mL conical vial using an analytical balance, and a volume of ethanol (200 proof) needed to make a suspension at 2.90 mg mL−1 was added using a micropipetter. This mixture was sonicated for ten minutes, with occasional manual shaking, forming a homogeneous suspension. Aliquots of this suspension (100.0 μL, 0.290 mg of APMS) were then dispensed into Eppendorf tubes (0.5 mL) and the ethanol was evaporated overnight in a 40 °C oven before finally drying under vacuum for two hours. Prior to the adsorption of siRNA, the vials were removed from the vacuum and the particles were irradiated with UV light (350 nm) for one hour to sterilize them and degrade any contaminating nucleases. In a sterile cell culture hood, RNase-free water was added (20 – 48 μL), the particles were allowed to be wet by the water for 10 min, and siRNA-AF647 was added from a 40.0 μM stock solution (2 – 30 μL) so that the total volume was 50.0 mL. The particles were wrapped in aluminum foil and rocked at room temperature. After 24 hours, the particles were centrifuged (8000 × g, 10 min), and aliquots (40.0 μL) were quickly removed via micropipetter to new, UV-irradiated Eppendorf tubes (0.5 mL). The supernatant was then diluted, as needed, and analysis was performed by UV/Vis spectroscopy. The amount of siRNA adsorbed was then calculated by the difference between that measured in the supernatant (ε650 = 563,000 M−1 cm−1, determined in-house) and the amount known to have been added from the stock solution. Each equilibrium concentration was measured in triplicate.
siRNA Release
To mimic conditions during cellular transfection, release was measured into the low serum growth medium used for the in vitro knockdown experiments described above. Particles were loaded with siRNA by first weighing them into Eppendorf tubes using a microbalance (2–3 mg), irradiating them with UV light (350 nm) for one hour, and adding appropriate volumes of siRNA (40.0 μM) and RNase-free water to maximally load the particles, given the adsorption isotherms measured above. After shaking for 24 hours, the loaded particles were isolated by centrifugation (2000 × g, 5 min, saving the supernatant to determine the amount adsorbed), washed quickly with deionized water, and resuspended in growth medium (2.00 mg mL−1). Three aliquots of this suspension (250. μL, 0.500 mg APMS) were divided into new Eppendorf tubes that contained pre-warmed medium (750. μL), creating three replicates. The tubes were wrapped in foil and suspended at 37 °C in a water bath. At various time points, aliquots (50.0 μL) of the suspension were removed and placed into UV-irradiated Eppendorf tubes (0.5 mL) that were immediately centrifuged (2000 × g, 1 min). The supernatant (10 – 40 μL) was then immediately removed via pipette for dilution, as needed, and analysis was performed by UV/Vis spectroscopy (ε650 = 563,000 M−1 cm−1).
Results and Discussion
A key requirement of transfection agents for siRNA is that the cargo be delivered to the cytosol for processing in the RISC complex. Existing particle-based transfection agents are nano-sized and are internalized in cells within endosomes. Thus, these agents must utilize the proton-sponge effect, the self-buffering capability of polyamine substituents,15,16 to deliver the siRNA to the cytosol. In contrast, we have previously shown that micron-sized APMS, when functionalized on its exterior surface with tetraethylene glycol chains, are not membrane-bound upon internalization,32 meaning that siRNA would be released directly to the cytosol without having to effect endosomal escape. We have also recently shown that pore modifications and pore size can greatly change the loading and release characteristics of DNA from mesoporous silica.43 Thus, we designed a systematic study of these factors on the uptake and release of siRNA from similar materials. Our goals were to optimize the nature of the pore modification, the degree of pore functionalization, and the pore size. In so doing, we also wanted to determine how much and how tightly siRNA bound to the particles, where the siRNA was bound within the particles, and how these factors affected the release of siRNA.
siRNA Adsorption
We selected diethylenetriamine (DETA) as the pore modification for siRNA adsorption and release experiments based on preliminary studies with several cationic groups (Figure S1 and Table S1 in the Supporting Information [SI]).43,49–52 DETA has a cationic charge at physiological pH and served to mediate the interactions of anionic siRNA to the silica substrate. All of the particles used in these studies were also functionalized with tetraethylene glycol on their exteriors to facilitate uptake into cells, as we have shown previously.32,47 As an siRNA payload, we used a commercially available, non-targeting siRNA labeled with the AlexaFluor®-647 dye, which allowed quantification by UV/Visible spectroscopy (see above for sequence). Langmuir adsorption isotherms were generated by measuring the amount of adsorbed siRNA as a function of equilibrium concentration after 24 hours. (See the Supporting Information for details on the curve fitting and the Langmuir equation). Release of siRNA from these materials was measured by suspending siRNA-loaded materials in low-serum growth medium for 24 hours to best replicate conditions used for later in vitro knockdown experiments. These preliminary experiments showed that DETA offered optimum adsorption and release characteristics for further studies, compared to the other cationic functional groups that we tested. In particular, although DETA released only 20 % of its adsorbed siRNA, the loading capacity (Bmax) was sufficiently large (24 μg per mg of APMS) that the total amount of released siRNA was the largest of the materials tested. Finally, these studies also indicated that the pore diameter had an influence on the amount of siRNA adsorbed; materials with 8 nm pores adsorbed more siRNA than materials with 4 nm pores, for a fixed amount of surface modification.
We then performed a systematic study of the effect of the amount of DETA modification on siRNA uptake, using APMS with 8 nm pores. To elucidate the effect of degree of functionalization, we modified particles with various amounts of DETA according to the method of Brühwiler.46 Briefly, APMS particles functionalized exclusively on their surfaces with tetraethylene glycol (“APMS-(s)TEG”) were stirred in a mixture of hexanes and dichloromethane with increasing amounts of 1-(3-trimethoxysilylpropyl)-diethylenetriamine (TMSP-DETA) for ten minutes (Scheme 1). After this time, the particles were filtered quickly without rinsing, allowed to air dry for 30 minutes, then cured at 80 °C overnight, yielding a series of particles referred to as APMS8-DETAx, where the subscript “8” indicates an average pore diameter of 8 nm and the subscript “x” indicates the mass percent of TMSP-DETA used in the functionalization solution compared to the mass of particles (x = 0.5, 1.5, 2.5, 7.5, and 15.0 %). Characterization of these particles using N2 physisorption is summarized in Table 1 and Figure S2. The degree of functionalization with DETA correlated monotonically with the amount of alkoxysilane in the modification procedure as evidenced by the decreased surface area and pore volume with increasing amounts of modification.
Scheme 1.
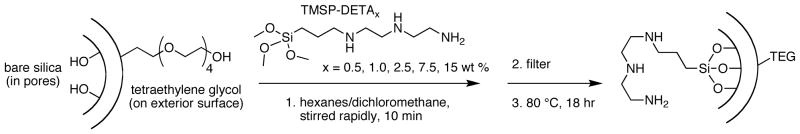
Modification of APMS-(s)TEG with varying amounts of TMSP-DETA.
Table 1.
Physical Properties of DETA-Modified APMS.
| DETA in loading solution (wt %) | dpore (nm) | Vpore (cm3 g−1) | SABET (m2 g−1) |
|---|---|---|---|
| - | 8 | 0.98 | 443 |
| 0.5 | 8 | 1.01 | 428 |
| 1.0 | 8 | 0.99 | 417 |
| 2.5 | 8 | 0.97 | 417 |
| 7.5 | 8 | 0.93 | 401 |
| 15 | 8 | 0.83 | 368 |
| - | 15 | 0.85 | 191 |
| 0.5 | 15 | 0.88 | 191 |
| 2.5 | 15 | 0.85 | 186 |
| 15 | 15 | 0.74 | 156 |
| - | 4 | 0.78 | 640 |
| 0.5 | 4 | 0.77 | 645 |
| 2.5 | 4 | 0.75 | 639 |
| 15 | 4 | 0.62 | 557 |
We then measured siRNA adsorption isotherms for these materials using the same procedure described above. The data in Figure S3 and Table 2 show that the adsorption characteristics were strongly dependent on the degree of DETA functionalization. Indeed, the values of Bmax increased monotonically from 13 to 36 μg mg−1 as the amount of DETA increased from 0.5 % to 15 %. This result demonstrates that the loading capacity of siRNA could easily be controlled by varying the density of amines on the pore surfaces of the silica particles, and as expected, the loading capacity increased with an increasing amount of modification. The dissociation constant Kd increased with increasing amounts of amine functionalization, indicating a weaker siRNA interaction with the modified material at higher loadings.53 This result could arise from increased siRNA-siRNA interactions as more material was adsorbed onto the surface. Despite the complexities of the molecular structure of the adsorbate and the functionalized silica pores, the Langmuir model of these samples revealed the modulation of adsorption properties as the amount of DETA on the surface increased.
Table 2.
Langmuir and Freundlich fitting parameters for the adsorption of siRNA into various APMS modified with DETA. Best fits (Langmuir vs. Freundlich) are in bold.
| dpore (nm) | DETA (wt %) | Langmuir
|
Freundlich
|
|||||
|---|---|---|---|---|---|---|---|---|
| Bmax (μg mg−1) | Bmax (μg m−2) | Kd (nM) | R2 | KF (μg m−2 M−A) | A | R2 | ||
|
|
|
|
||||||
| 4 | 0.5 | 14 ± 0.6 | 21 ± 1 | 41 ± 6 | .926 | 624 ± 260 | 0.239 | .862 |
| 2.5 | 14 ± 0.4 | 20 ± 0.4 | 20 ± 2 | .960 | 280 ± 80 | 0.183 | .948 | |
| 15 | 18 ± 0.4 | 32 ± 1 | 40 ± 3 | .973 | 570 ± 210 | 0.208 | .831 | |
| 8 | 0.5 | 13 ± 0.3 | 31 ± 1 | 19 ± 2 | .960 | 440 ± 90 | 0.191 | .941 |
| 2.5 | 22 ± 0.6 | 53 ± 2 | 87 ± 9 | .978 | 3600 ± 1500 | 0.309 | .915 | |
| 15 | 36 ± 2 | 96 ± 5 | 220 ± 20 | .987 | 340,000 ± 15,000 | 0.583 | .978 | |
| 15 | 0.5 | 13 ± 0.5 | 67 ± 2 | 41 ± 7 | .891 | 950 ± 200 | 0.196 | .928 |
| 2.5 | 20 ± 1 | 108 ± 4 | 100 ± 10 | .939 | 2300 ± 500 | 0.231 | .959 | |
| 15 | 35 ± 1 | 230 ± 10 | 270 ± 30 | .965 | 35,000 ± 4000 | 0.384 | .994 | |
|
|
|
|
||||||
Having characterized the adsorption of siRNA into DETA-functionalized APMS with 8.0 nm pores, we explored the interplay of pore diameter and extent of DETA functionalization by synthesizing two additional series of particles and examining their uptake of siRNA. Specifically, we prepared batches of APMS particles with 4 nm pores and 15 nm pores using techniques developed in our laboratory34,35 and modified some of each material with 0.5 %, 2.5 %, and 15 % TMSP-DETA, as described above. Again, N2 physisorption experiments demonstrated monotonically increasing functionalization by the DETA polyamine with increasing amounts of silane used in the modification procedure (Table 1). We then repeated the adsorption experiments with these new materials (Figure 1 and Table 2). The particles with 15 nm pores and varying amounts of DETA, designated APMS15-DETAx, displayed trends consistent with the 8 nm pore materials. Specifically, the loading capacity of siRNA increased monotonically with increasing amounts of amine functionalization, from 67 to 230 μg m−2. These values, normalized to the specific surface area of each particle type, were appreciably larger than the particles with 8 nm pores. However, the relatively small surface area of the large-pore APMS15-DETAx (156–191 m2 g−1) compared to the medium-pore APMS8-DETAx (368–428 m2 g−1) resulted in values of Bmax that were not substantially different between the two pore sizes on a mass basis – 13 to 35 μg mg−1 for materials with 15 nm pores, compared to 13 to 36 μg mg−1 for materials with 8 nm. Values of Kd were also similar for both of these materials. The particles with 4 nm pores showed an interesting divergence from the adsorption characteristics observed with the pore diameters of 8 nm and 15 nm. These materials, designated APMS4-DETAx, displayed a much weaker dependence of both Bmax and Kd on the degree of functionalization (Figure 1A). Indeed, the maximum specific adsorption of siRNA in APMS4-DETAx spanned the narrow range of 21 to 32 μg m−2 (14 to 18 μg mg−1) even with the same wide range of DETA modification as in the other particles. Also, the values of Kd, 20 to 41 nM, were essentially not a function of DETA modification in the range studied here.
Figure 1.
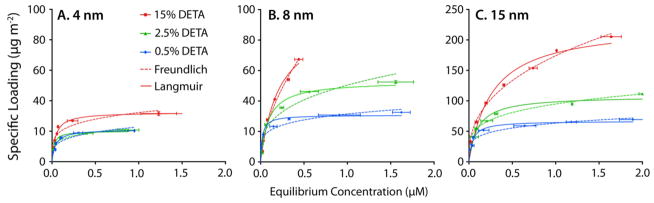
Adsorption of siRNA into APMS with pore diameters of either 4 (A), 8 nm (B), or 15 nm (C) and modified with varying amounts of DETA, as indicated in each plot. Data are fitted to Langmuir (solid lines) and Freundlich (dashed lines) isotherms.
In addition to the Langmuir adsorption isotherms, we also calculated parameters for the Freundlich model of adsorption to gain further insight into the mode(s) of adsorption occurring in these systems. (See the Supporting Information for the Freundlich equation). The Freundlich model is a semiempirical model that, unlike the Langmuir model, lacks a saturation loading capacity and can account for both the formation of multilayers and for heterogeneous binding sites on the surface.43,53 The Langmuir and Freundlich parameters for the adsorption of siRNA into APMS as a function of pore size and % DETA modification are tabulated in Table 2. The best model for each material was determined by the largest value of R2, and the parameters for the corresponding model have been shown in bold-faced type in the table. Interestingly, the Langmuir model was superior for all cases of the smaller-sized pores (4 nm and 8 nm), suggesting that siRNA adsorbed in monolayers with pores of this size range. On the other hand, the Freundlich model better fit the adsorption isotherms for all of the 15 nm pore materials, indicating that multilayer adsorption was occurring. Because the pore-immobilized DETA molecules could interact with siRNA only near the pore surfaces, they could not mitigate repulsions between the oligonucleotides in the middle of the pores, no matter how large the degree of functionalization, causing a decrease in binding strength with increasing loading capacity.
Unlike the larger-pore materials, we found that the adsorption characteristics were essentially not a function of degree of functionalization in the particles with 4 nm pores. Because the siRNA had a diameter of ~3 nm,54–56 these results suggested that the loading capacity and the binding strength of the siRNA was geometrically limited to only a one-molecule-deep monolayer. Apparently, even the least-modified surface provided a sufficiently strong interaction to bind nearly a pore-filling (and thus maximal) amount of siRNA. Then, even with increasing DETA, the pores could accommodate no more adsorbate, resulting in no additional saturation loading capacity and therefore none of the additional adsorbate-adsorbate repulsions that had increased the values of Kd in the larger-pore materials.
The results of the adsorption isotherm curve fitting further suggested that the pore diameters of these materials caused important differences in their adsorptive characteristics. Adsorption into the 4 nm and 8 nm pores was best described by the monolayer model, while 15 nm pores were best described by the multilayer model. This result is nicely explained by the work of Coppens and co-workers,57 who used geometric considerations to determine the maximum number of adsorbate molecules that can fit in a pore of a given diameter. The amount of material that can adsorb in the pores per mass of particle, Γinternal, is equal to
| (1) |
where M is the molar mass of the adsorbate, δ is the largest dimension of the adsorbate, NA is Avagadro’s number, Vt is the total pore volume of the mesoporous material, Ap is the average cross-sectional pore area of the pores, and Npore is the number of adsorbate molecules that can be accommodated within the pore, given by
| (2) |
where De is the average diameter of the two shorter dimensions of the adsorbate, Dp is the diameter of the mesopores, and n is the number of layers formed by the adsorbate. Because of the large size of APMS (> 1 μm), almost all of the surface area is from the pores, so we have ignored the contribution from adsorption to the exterior of the particles.
We used Equations 1 and 2 and assumptions about the dimensions of our siRNA (20 base pairs, 6 nm long × 3 nm in diameter)54–56,58 to calculate that an 8 nm pore could accommodate only a 4-molecule monolayer of oligonucleotides (Equation 1 and Figure 2, left), resulting in monolayer adsorption characteristics modeled well by the Langmuir equation. Conversely, 15 nm pores could accommodate a 12-molecule monolayer with an additional sextet filling the pores, resulting in multilayer adsorption characteristics better fit by the Freundlich model (Figure 2, middle). The adsorption would have plateaued at even higher equilibrium concentrations of siRNA as the pores eventually filled, but this apparently had not yet occurred in the range of equilibrium concentrations studied here. Alternatively, the arrangement of siRNA could have been different from that predicted by Coppens’s model since our adsorbates were not at their isoelectric points. For example, repulsions between the siRNA molecules could have resulted in partial end-on binding to the pore walls at high DETA functionalization in materials with 15 nm pores. Assuming half end-on and half edge-on binding (for an average “diameter” of 4 nm), the calculation predicts an 8-molecule monolayer (Figure 2, right). Interestingly, this binding arrangement would result in a per-surface-area loading capacity twice that of the 4-molecule monolayer that occurred in the 8 nm pores, which is approximately what we observed (230 μg m−2 vs. 96 μg m−2). This finding, together with the good fits from the Freundlich model, suggested that some combination of adsorption processes (monolayer and multilayer) occurred in the material with 15 nm pores. We note that such combinations of adsorption modes were possible only in a material with pores larger than ~12 nm because the oligonucleotides were roughly 6 nm long, and such arrangements require pore diameters greater than or equal to two lengths of siRNA.
Figure 2.
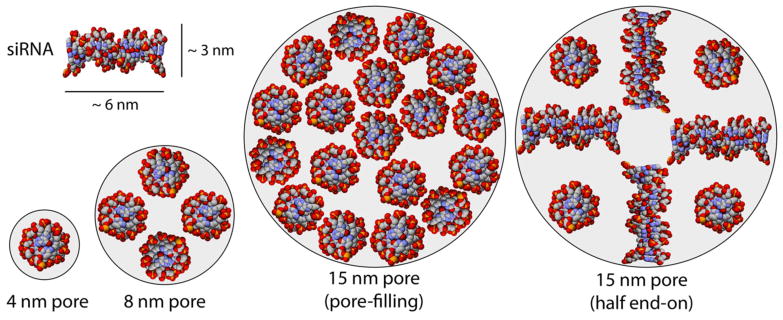
Examples of possible siRNA binding modes in silica pores with diameters of 4, 8, or 15 nm.
In addition to developing the adsorption isotherms of siRNA into APMS, we used confocal microscopy to determine the spatial distribution of the dye-labeled (AlexaFluor-647) siRNA within the particles as a function of loading. Following siRNA loading as described above, particles were washed twice with water to remove adventitious siRNA and fixed to slides for confocal microscopy. For each adsorption experiment, we imaged particles that were only slightly loaded with siRNA (i.e. those to the far left of an adsorption isotherm) and particles that were fully loaded with siRNA (to the far right) after 24 hr of equilibration. The images show that for all degrees of modification and all pore sizes, the appearance of rings of fluorescence at low equilibrium concentrations of siRNA suggested that the adsorption occurred first at the periphery of the particles (Figure 3; panels A, C, and E). When the equilibrium concentration of siRNA was increased to levels that achieved maximal loading, the ring-like appearance was replaced by solid disks of fluorescence, indicating that the siRNA had penetrated to pores throughout the spherical microparticles (Figure 3, panels B, D, and F). These observations imply that steric crowding near the mouth of the pores caused by the initial adsorption events blocked subsequent uptake at sites deeper within the particles in the absence of excess siRNA (i.e. at low equilibrium concentration). Then, when greater amounts of siRNA were available, adsorption became favorable throughout the particle. Alternatively, it is possible that the distribution of amine groups was not homogeneous throughout the particles. For instance, the most densely functionalized regions could have been the periphery of the particles, resulting in the strongest (and therefore, the initial) adsorption there. Regardless of the exact details of the adsorption process, these confocal images show that siRNA was adsorbed throughout the porous network of APMS particles. This result is in contrast to other studies in which siRNA was adsorbed only to the outside of the particles,22–24 a difference that is potentially important for the protection of the siRNA payload from RNAses in vivo. Finally, we note that the APMS particles remained well-dispersed in all the confocal micrographs, indicating that loading with siRNA had not caused irreversible aggregation of the particles.
Figure 3.
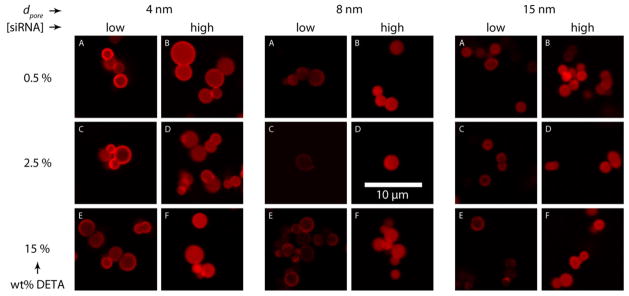
Confocal microscopy of APMS particles loaded with fluorescently labeled siRNA. The figure compares the loading of siRNA at both low and high siRNA equilibrium concentrations for materials at several pore diameters, each with several levels of DETA surface modifications.
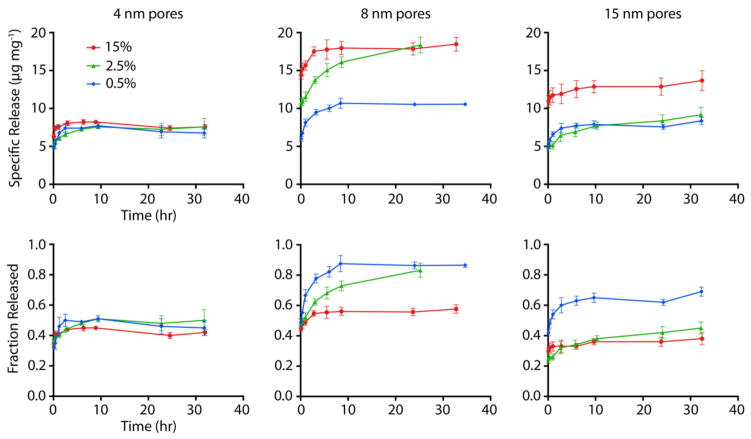
To examine the release characteristics of the siRNA from DETA-functionalized APMS, the particles were suspended in aqueous solution of siRNA for 24 h using enough siRNA to ensure maximum uptake, given the adsorption experiments above. Release was then measured by stirring the siRNA-loaded particles at 37 °C in cell culture medium used for in vitro transfection experiments; the release was quantified by UV/Visible spectroscopy using the AlexaFluor-647 tag on the siRNA. We found that much of the adsorbed siRNA could be released, with the pore diameter and degree of functionalization again strongly affecting the release characteristics (Figure 4 and Table 3). Particles with 4 nm pores released essentially identical amounts of siRNA, between 6.5 and 7.5 μg mg−1, regardless of the extent of DETA surface modification. The fraction of loaded siRNA that was released from these materials showed little variation across the series, changing only from 0.45 to 0.51. In contrast, materials with larger pores (both 8 and 15 nm) showed a marked change in specific release as the amount of DETA surface modification increased, nearly doubling in the amount of siRNA released. Despite increases in the absolute amounts of released siRNA, the released fraction decreased monotonically for these materials as the amount of DETA increased (0.87 to 0.59 for 8 nm materials and 0.69 to 0.38 for 15 nm materials). Thus, although the more heavily modified particles could adsorb more siRNA, their delivery efficiency decreased.
Figure 4.
Release kinetics of siRNA-AlexaFluor647 from APMS as functions of pore size (4, 8, and 15 nm) and degree of functionalization with DETA (0.5, 2.5, and 15 % as described above). The amount of polyamine modification had relatively little impact on the release characteristics of the materials with 4 nm pores (left). Conversely, the amount of modification had a substantial effect for materials with 8 and 15 nm pores.
Table 3.
Summary of data for the release of siRNA from various APMS particles.
| dpore (nm) | DETA (wt %) | Maximum siRNA Released
|
|
|---|---|---|---|
| Total Mass (μg mg−1) | Fraction of Adsorbed Amount | ||
| 4 | 0.5 | 7.7 | 0.51 |
| 2.5 | 7.6 | 0.51 | |
| 15 | 8.2 | 0.45 | |
| 8 | 0.5 | 11 | 0.88 |
| 2.5 | 18 | 0.83 | |
| 15 | 19 | 0.58 | |
| 15 | 0.5 | 8.4 | 0.69 |
| 2.5 | 9.2 | 0.45 | |
| 15 | 14 | 0.38 | |
|
|
|||
Materials with the larger pore diameters released the smallest fraction of siRNA at intermediate and high degrees of functionalization, compared to all other materials. This could occur because the larger pores allowed the rearrangement of adsorbed siRNA into geometries that mitigated molecular repulsions between siRNA molecules through increased distance or the influx of ions such as Mg+2 from the release medium, while still retaining much of the adsorbed siRNA. As shown in Figure 2, significant adsorbate repulsions are induced in 8 nm pores as compared to 15 nm pores. Indeed, the ideal combination of pore diameter and degree of functionalization appeared to be 8 nm pores with 2.5 % DETA functionalization, which released the same absolute amount of siRNA as the most heavily functionalized materials of any pore diameter (~ 19 μg mg−1) given that this amount represented a large fraction of the adsorbed material (0.83).
The shapes of the release curves show that all particle types exhibited a significant “burst” release of siRNA less than five minutes after immersion in cell culture medium. When the particles were washed with water alone, this burst release did not occur, as measured by the method outlined above, in control experiments. This finding suggests that either one or more of the many components of the culture medium exhibited preferential adsorption to the particles, or that the siRNA itself was drawn out of the particles due to energetically favorable interactions with one of the components. The speed of this initial release indicated that diffusion could occur with very little kinetic barrier, in contrast to other cases of release from mesopores into less complex solvent systems.43,59,60 Alternatively, the burst release could correspond to loosely bound siRNA due to multilayer adsorption or from release at the pore entrances, where little diffusion through pores would be required. This result indicates that controlled release strategies, such as reduction-sensitive disulfide linkers that have found success in other materials,29,61,62 may be required to avoid premature release if more potent siRNA delivery within cells is desired.
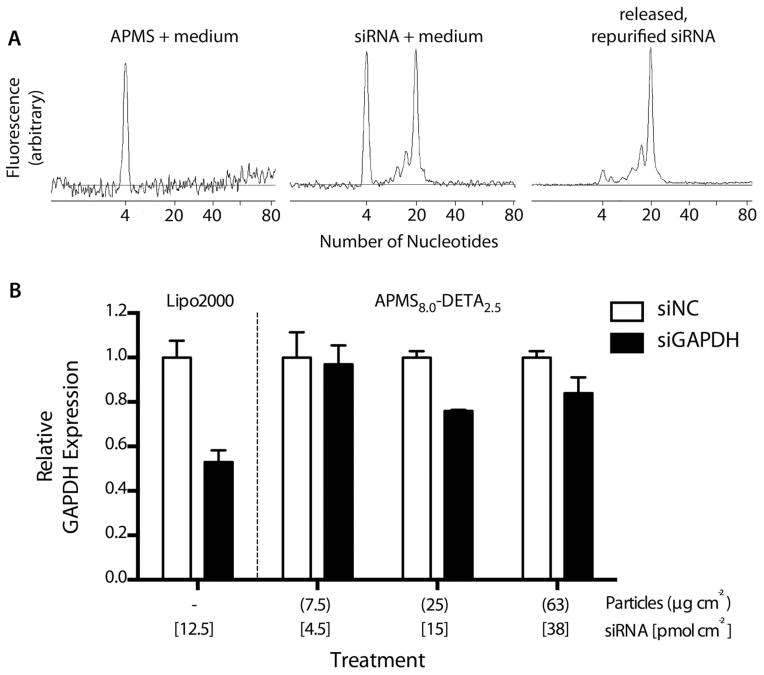
To demonstrate that siRNA was released from APMS-DETA intact and functional, we performed capillary electrophoresis (CE) and in vitro transfection experiments using quantitative real-time polymerase-chain reaction (qRT-PCR). Given the release data above, we chose the material with 8 nm pores and 2.5% DETA modification as the best compromise between maximum loading and release amounts and kinetics. We selected glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as our gene of interest because it is commonly used as a knockdown control and a validated, positive-control siRNA was commercially available. For the CE experiments, particles were loaded with siRNA from water as described above and released into growth medium but without removing aliquots. After release into medium for 24 hr, the siRNA was isolated by solid-phase extraction and analyzed by CE. The electrophoretogram of the siRNA that was loaded into APMS, released, and purified was nearly identical to the stock siRNA mixed with the growth medium (Figure 5A), showing that the GAPDH siRNA was released from APMS-DETA as an intact oligonucleotide. We then used these particles as a transfection agent for in vitro experiments using small airway epithelial cells (SAEC).44 qRT-PCR was used to measure mRNA levels of the GAPDH gene, and a validated negative control siRNA was used as control. Particles were loaded with one of the siRNA constructs for 24 hr, then isolated by centrifugation, washed once with water, resuspended in PBS, and administered to SAEC cells at varying doses that we have used previously to deliver small-molecule drugs and DNA plasmids.32,63 The qRT-PCR results showed that administration of siRNA-loaded APMS-DETA particles led to a GAPDH mRNA knockdown to 76 % of the negative control after 46 hr (Figure 5B). For comparison, using the commercially-available polymeric transfection agent Lipofectamine 2000® resulted in a knockdown to 53 % of the negative control GAPDH expression, roughly twice as much knockdown as the particles.
Figure 5.
siRNA oligonucleotides are released intact and retain their functionality in vitro as demonstrated by capillary electrophoresis (A) and real-time, quantitative PCR (B). The electrophoretograms show that siRNA that was loaded into APMS particles, released from the particles, and repurified (top right) is essentially identical to the same siRNA from the stock solution added directly to growth medium, simulating the release environment within a biological system (top middle). Also, APMS particles were able to deliver functional siRNA to the interiors of cells as shown by a decrease in the expression of GAPDH, the protein whose gene was targeted by our siRNA sequence (bottom).
Conclusions
We have presented the first systematic study of the effects of pore size and pore modification on the adsorption and release of siRNA from acid-prepared mesoporous silica (APMS). We found that a short polyamine, diethylenetriamine (DETA), was the best chemical modification of the pores for achieving both the adsorption and release of large amounts of siRNA. Adsorption isotherms demonstrated that the degree of functionalization with DETA can cause drastic changes to the loading capacity and binding strength of siRNA to silica with relatively large pores (8 nm and larger), but degree of functionalization has a much weaker effect in narrow pores (4 nm) that could accommodate only one siRNA molecule. Moreover, the adsorption data suggested that multilayer adsorption could occur in materials with large pores (15 nm). Release experiments into cell culture medium showed that intermediate pore sizes and intermediate degrees of functionalization resulted in the best compromise between maximizing loading (from strong adsorption) and maximizing release. Capillary electrophoresis and in vitro transfection experiments demonstrated that the siRNA was released intact and that APMS-DETA could function as a transfection agent.
Supplementary Material
Acknowledgments
This project was funded by the National Institutes of Health under grant numbers T32ES0071 and 1R41-CA126155-01A1 and by the Mesothelioma Applied Research Foundation. The authors would like to thank Prof. Arti Shukla for her assistance with qRT-PCR measurements and the Vermont Cancer Center Advanced Genome Technologies Core facility.
Footnotes
Supporting Information Available
Additional siRNA adsorption plots, N2 physisorption isotherms, pore size distribution plots, tabulated characterization information, and mathematical models. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007 - an update. J Gene Med. 2007;9:833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 3.Reischl D, Zimmer A. Drug delivery of siRNA therapeutics: potentials and limits of nanosystems. Nanomed-Nanotechnol Biol Med. 2009;5:8–20. doi: 10.1016/j.nano.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 5.Persengiev SP, Zhu XC, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA-Publ RNA Soc. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BRG. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 7.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA-Publ RNA Soc. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, Polisky BA, Zinnen S. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 9.Gao S, Dagnaes-Hansen F, Nielsen EJB, Wengel J, Besenbacher F, Howard KA, Kjems J. The Effect of Chemical Modification and Nanoparticle Formulation on Stability and Biodistribution of siRNA in Mice. Mol Ther. 2009;17:1225–1233. doi: 10.1038/mt.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heller R, Jaroszeski M, Atkin A, Moradpour D, Gilbert R, Wands J, Nicolau C. In vivo gene electroinjection and expression in rat liver. FEBS Lett. 1996;389:225–228. doi: 10.1016/0014-5793(96)00590-x. [DOI] [PubMed] [Google Scholar]
- 11.Golzio M, Mazzolini L, Ledoux A, Paganin A, Izard M, Hellaudais L, Bieth A, Pillaire MJ, Cazaux C, Hoffmann JS, Couderc B, Teissie J. In vivo gene silencing in solid tumors by targeted electrically mediated siRNA delivery. Gene Ther. 2007;14:752–759. doi: 10.1038/sj.gt.3302920. [DOI] [PubMed] [Google Scholar]
- 12.Walther W, Stein U. Viral vectors for gene transfer - A review of their use in the treatment of human diseases. Drugs. 2000;60:249–271. doi: 10.2165/00003495-200060020-00002. [DOI] [PubMed] [Google Scholar]
- 13.Uchida H, Tanaka T, Sasaki K, Kato K, Dehari H, Ito Y, Kobune M, Miyagishi M, Taira K, Tahara H, Hamada H. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther. 2004;10:162–171. doi: 10.1016/j.ymthe.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Ling B, Zhou Y, Feng DQ, Shen GD, Gao T, Shi YY, Wei HM, Tian ZG. Down-regulation of EDAG expression by retrovirus-mediated small interfering RNA inhibits the growth and IL-8 production of leukemia cells. Oncol Rep. 2007;18:659–664. [PubMed] [Google Scholar]
- 15.Grayson ACR, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 16.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 17.Davis ME, EZJ, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1071. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, Dames S, Loffler K, Fechtner M, Arnold W, Giese K, Klippel A, Kaufmann J. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 20.Villares GJ, Zigler M, Wang H, Melnikova VO, Wu H, Friedman R, Leslie MC, Vivas-Mejia PE, Lopez-Berestein G, Sood AK, Bar-Eli M. Targeting Melanoma Growth and Metastasis with Systemic Delivery of Liposome-Incorporated Protease-Activated Receptor-1 Small Interfering RNA. Cancer Res. 2008;68:9078–9086. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Saad M, Pakunlu RI, Khandare JJ, Garbuzenko OB, Vetcher AA, Soldatenkov VA, Pozharov VP, Minko T. Nonviral nanoscale-based delivery of antisense oligonucleotides targeted to hypoxia-inducible factor 1 alpha enhances the efficacy of chemotherapy in drug-resistant tumor. Clin Cancer Res. 2008;14:3607–3616. doi: 10.1158/1078-0432.CCR-07-2020. [DOI] [PubMed] [Google Scholar]
- 22.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VSY. A polyamidoamine dendrimer-capped mesoporous silica nanosphere-based gene transfection reagent. J Am Chem Soc. 2004;126:13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 23.Chen AM, Zhang M, Wei DG, Stueber D, Taratula O, Minko T, He HX. Co-delivery of Doxorubicin and Bcl-2 siRNA by Mesoporous Silica Nanoparticles Enhances the Efficacy of Chemotherapy in Multidrug-Resistant Cancer Cells. Small. 2009;5:2673–2677. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia TA, Kovochich M, Liong M, Meng H, Kabehie S, George S, Zink JI, Nel AE. Polyethyleneimine Coating Enhances the Cellular Uptake of Mesoporous Silica Nanoparticles and Allows Safe Delivery of siRNA and DNA Constructs. ACS Nano. 2009;3:3273–3286. doi: 10.1021/nn900918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashley CE, Carnes EC, Epler KE, Padilla DP, Phillips GK, Castillo RE, Wilkinson DC, Wilkinson BS, Burgard CA, Kalinich RM, Townson JL, Chackerian B, Willman CL, Peabody DS, Wharton W, Brinker CJ. Delivery of Small Interfering RNA by Peptide-Targeted Mesoporous Silica Nanoparticle-Supported Lipid Bilayers. ACS Nano. 2012;6:2174–2188. doi: 10.1021/nn204102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartono SB, Gu W, Kleitz F, Liu J, He L, Middelberg APJ, Yu C, Lu GQ, Qiao SZ. Poly L-Lysine Functionalized Large Pore Cubic Mesostructured Silica Nanoparticles as Biocompatible Carriers for Gene Delivery. 2012;6:2104–2117. doi: 10.1021/nn2039643. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Chen Y, Wang M, Ma Y, Xia W, Gu H. A mesoporous silica nanoparticle - PEI - Fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials. 2013;34:1391–1401. doi: 10.1016/j.biomaterials.2012.10.072. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Xie QR, Zhang J, Xia W, Gu H. The packaging of siRNA within the mesoporous structure of silica nanoparticles. Biomaterials. 2011;32:9546–9556. doi: 10.1016/j.biomaterials.2011.08.068. [DOI] [PubMed] [Google Scholar]
- 29.Lin D, Cheng Q, Jiang Q, Huang Y, Yang Z, Han S, Zhao Y, Guo S, Liang Z, Dong A. Intracellular cleavable poly(2-dimethylaminoethyl methacrylate) functionalized mesoporous silica nanoparticles for efficient siRNA delivery in vitro and in vivo. Nanoscale. 2013;5:4291–4301. doi: 10.1039/c3nr00294b. [DOI] [PubMed] [Google Scholar]
- 30.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J Am Chem Soc. 2009;131:2072. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Lee K, Moon SH, Lee Y, Park TG, Cheon J. All-in-One Target-Cell-Specific Magnetic Nanoparticles for Simultaneous Molecular Imaging and siRNA Delivery. Angew Chem-Int Edit. 2009;48:4174–4179. doi: 10.1002/anie.200805998. [DOI] [PubMed] [Google Scholar]
- 32.Blumen SR, Cheng K, Ramos-Nino ME, Taatjes DJ, Weiss DJ, Landry CC, Mossman BT. Unique uptake of acid-prepared mesoporous spheres by lung epithelial and mesothelioma cells. Am J Respir Cell Mol Biol. 2007;36:333–342. doi: 10.1165/rcmb.2006-0319OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nassivera T, Eklund AG, Landry CC. Size-exclusion chromatography of low-molecular-mass polymers using mesoporous silica. J Chromatogr A. 2002;973:97–101. doi: 10.1016/s0021-9673(02)01200-1. [DOI] [PubMed] [Google Scholar]
- 34.Gallis KW, Araujo JT, Duff KJ, Moore JG, Landry CC. The use of mesoporous silica in liquid chromatography. Adv Mater. 1999;11:1452–1455. [Google Scholar]
- 35.Gallis KW, Landry CC. 6,334,988. United States Patent. 2002
- 36.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 37.Oberdorster G, Oberdorster E, Oberdorster J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buzea C, Pacheco II, Robbie K. Nanomaterials and nanoparticles: Sources and toxicity. Biointerphases. 2007;2:MR17–MR71. doi: 10.1116/1.2815690. [DOI] [PubMed] [Google Scholar]
- 39.Casals E, Vazquez-Campos S, Bastus NG, Puntes V. Distribution and potential toxicity of engineered inorganic nanoparticles and carbon nanostructures in biological systems. Trac-Trends Anal Chem. 2008;27:672–683. [Google Scholar]
- 40.Hudson SP, Padera RF, Langer R, Kohane DS. The biocompatibility of mesoporous silicates. Biomaterials. 2008;29:4045–4055. doi: 10.1016/j.biomaterials.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzpatrick JAJ, Andreko SK, Ernst LA, Waggoner AS, Ballou B, Bruchez MP. Long-term Persistence and Spectral Blue Shifting of Quantum Dots in Vivo. Nano Lett. 2009;9:2736–2741. doi: 10.1021/nl901534q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewinski N, Colvin V, Drezek R. Cytotoxicity of nanoparticles. Small. 2008;4:26–49. doi: 10.1002/smll.200700595. [DOI] [PubMed] [Google Scholar]
- 43.Solberg SM, Landry CC. Adsorption of DNA into mesoporous silica. J Phys Chem B. 2006;110:15261–15268. doi: 10.1021/jp061691+. [DOI] [PubMed] [Google Scholar]
- 44.Piao CQ, Liu L, Zhao YL, Balajee AS, Suzuki M, Hei TK. Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis. 2005;26:725–731. doi: 10.1093/carcin/bgi016. [DOI] [PubMed] [Google Scholar]
- 45.Shukla A, Hillegass JM, MacPherson MB, Beuschel SL, Vacek PM, Pass HI, Carbone M, Testa JR, Mossman BT. Blocking of ERK1 and ERK2 sensitizes human mesothelioma cells to doxorubicin. Mol Cancer. 2010;9:13. doi: 10.1186/1476-4598-9-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gartmann N, Brühwiler D. Controlling and Imaging the Functional-Group Distribution on Mesoporous Silica. Angew Chem-Int Edit. 2009;48:6354–6356. doi: 10.1002/anie.200902436. [DOI] [PubMed] [Google Scholar]
- 47.Cheng K, Blumen SR, MacPherson MB, Steinbacher JL, Mossman BT, Landry CC. Enhanced Uptake of Porous Silica Microparticles by Bifunctional Surface Modification with a Targeting Antibody and a Biocompatible Polymer. ACS Appl Mater Interfaces. 2010;2:2489–2495. doi: 10.1021/am100530t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinbacher JL, Lathrop SA, Cheng K, Hillegass JM, Butnor K, Kauppinen RA, Mossman BT, Landry CC. Gd-Labeled Microparticles in MRI: In vivo Imaging of Microparticles After Intraperitoneal Injection. Small. 2010;6:2678–2682. doi: 10.1002/smll.201001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leal O, Anderson DL, Bowman RG, Basolo F, Burwell RL., Jr Reversible adsorption of oxygen on silica gel modified by imidazole-attached iron tetraphenylporphyrin. J Am Chem Soc. 1975;97:5125–5129. [Google Scholar]
- 50.Udayakumar S, Lee MK, Shim HL, Park SW, Park DW. Imidazolium derivatives functionalized MCM-41 for catalytic conversion of carbon dioxide to cyclic carbonate. Catal Commun. 2009;10:659–664. [Google Scholar]
- 51.Gao W, Xiao Z, Radovic-Moreno A, Shi J, Langer R, Farokhzad OC. Progress in siRNA Delivery Using Multifunctional Nanoparticles. In: Sioud M, editor. RNA Therapeutics. Springer; 2010. p. 629. [DOI] [PubMed] [Google Scholar]
- 52.Uchida H, Miyata K, Oba M, Ishii T, Suma T, Itaka K, Nishiyama N, Kataoka K. Odd-Even Effect of Repeating Aminoethylene Units in the Side Chain of N-Substituted Polyaspartamides on Gene Transfection Profiles. J Am Chem Soc. 2011;133:15524–15532. doi: 10.1021/ja204466y. [DOI] [PubMed] [Google Scholar]
- 53.Masel RI. Principles of Adsorption and Reaction on Solid Surfaces. Wiley-Interscience; New York, NY: 1996. p. 824. [Google Scholar]
- 54.Desigaux L, Sainlos M, Lambert O, Chevre R, Letrou-Bonneval E, Vigneron JP, Lehn P, Lehn JM, Pitard B. Self-Assembled lamellar complexes of siRNA with lipidic aminoglycoside derivatives promote efficient siRNA delivery and interference. P Natl Acad Sci USA. 2007;104:16534–16539. doi: 10.1073/pnas.0707431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:6968. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svintradze DV, Mrevlishvili GM. Fiber molecular model of atelecollagen-small interfering RNA (siRNA) complex. Int J Biol Macromol. 2005;37:283–286. doi: 10.1016/j.ijbiomac.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Sang LC, Vinu A, Coppens MO. General Description of the Adsorption of Proteins at Their Iso-electic Point in Nanoporous Materials. 2011;27:13828–13837. doi: 10.1021/la202907f. [DOI] [PubMed] [Google Scholar]
- 58.Bloomfield VA, Crothers DM, Tinoco I., Jr . Nucleic Acids: Structures, Properties, and Functions. Unversity Science Books; Sausalito, CA: 2000. [Google Scholar]
- 59.Cheng K, Landry CC. Diffusion-based deprotection in mesoporous materials: A strategy for differential functionalization of porous silica particles. J Am Chem Soc. 2007;129:9674–9685. doi: 10.1021/ja070598b. [DOI] [PubMed] [Google Scholar]
- 60.Lee CH, Lo LW, Mou CY, Yang CS. Synthesis and Characterization of Positive-Charge Functionalized Mesoporous Silica Nanoparticles for Oral Drug Delivery of an Anti-Inflammatory Drug. Adv Funct Mater. 2008;18:3283–3292. [Google Scholar]
- 61.Dunn SS, Tian S, Blake S, Wang J, Galloway AL, Murphy A, Pohlhaus PD, Rolland JP, Napier ME, DeSimone JM. Reductively Responsize siRNA-Conjugated Hydrogel Nanoparticles for Gene Silencing. J Am Chem Soc. 2012;134:7423–7430. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang G, Liu J, Qizhi Yang, Zhuo R, Jiang X. Disulfide-Containing Brushed Polyethylenimine Derivative Synthesized by Click Chemistry for Nonviral Gene Delivery. Bionconjugate Chem. 2012;23:1290–1299. doi: 10.1021/bc300133r. [DOI] [PubMed] [Google Scholar]
- 63.Hillegass JM, Blumen SR, Cheng K, MacPherson MB, Alexeeva V, Lathrop SA, Beuschel SL, Steinbacher JL, Butnor KJ, Ramos-Nino ME, Shukla A, James TA, Weiss DJ, Taatjes DJ, Pass HI, Carbone M, Landry CC, Mossman BT. Increased efficacy of doxorubicin delivered in multifunctional microparticles for mesothelioma therapy. Int J Cancer. 2011;129:233–244. doi: 10.1002/ijc.25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.